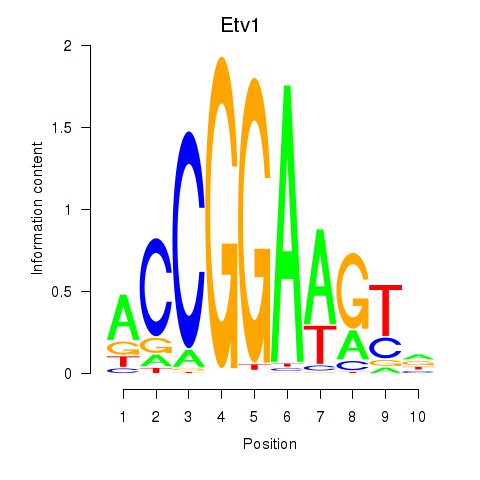
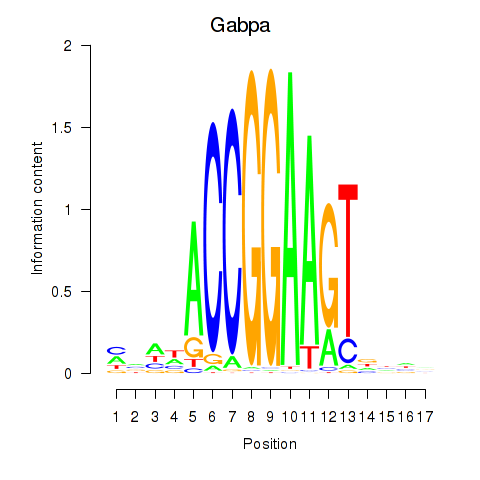
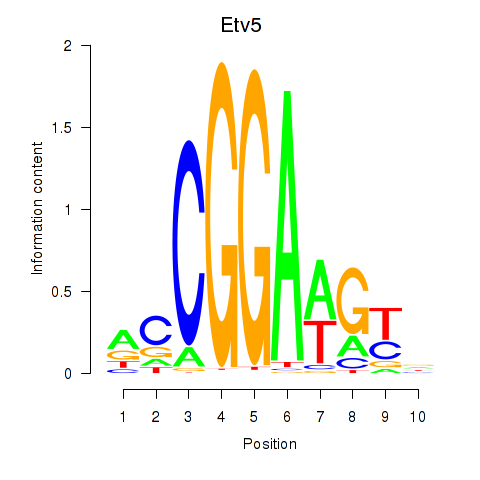
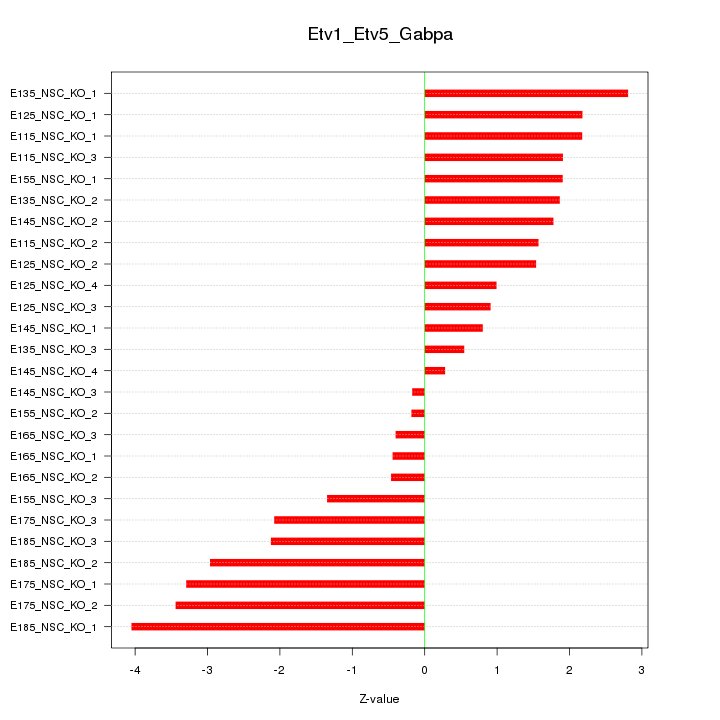
| 3.1 |
12.5 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 2.6 |
7.9 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.4 |
7.3 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 2.2 |
6.7 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 1.8 |
5.4 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 1.8 |
5.4 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.8 |
5.3 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 1.6 |
8.1 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.6 |
4.8 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 1.6 |
4.7 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 1.5 |
6.1 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 1.5 |
3.0 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 1.5 |
7.3 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 1.3 |
3.9 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 1.3 |
3.9 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 1.3 |
2.5 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 1.3 |
5.0 |
GO:0006203 |
dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 1.2 |
3.7 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 1.2 |
3.6 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 1.2 |
3.6 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 1.2 |
5.9 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 1.2 |
3.5 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 1.2 |
9.3 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 1.2 |
1.2 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 1.1 |
5.6 |
GO:0071038 |
nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) |
| 1.1 |
4.4 |
GO:1903899 |
positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.1 |
3.3 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 1.1 |
2.2 |
GO:0009146 |
purine nucleoside triphosphate catabolic process(GO:0009146) |
| 1.1 |
5.3 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.0 |
4.0 |
GO:0035624 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) response to high density lipoprotein particle(GO:0055099) |
| 1.0 |
4.0 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 1.0 |
1.0 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 1.0 |
10.5 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.9 |
13.3 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.9 |
7.6 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.9 |
7.4 |
GO:1901977 |
negative regulation of cell cycle checkpoint(GO:1901977) |
| 0.9 |
16.6 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.9 |
5.5 |
GO:0000447 |
endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.9 |
3.7 |
GO:1904430 |
negative regulation of t-circle formation(GO:1904430) |
| 0.9 |
4.5 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.9 |
0.9 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.9 |
3.5 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.9 |
4.3 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.9 |
2.6 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.8 |
4.2 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.8 |
2.5 |
GO:0006296 |
nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.8 |
2.5 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.8 |
3.2 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.8 |
3.2 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.8 |
0.8 |
GO:0031990 |
mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.8 |
2.3 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.8 |
3.0 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.7 |
3.7 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.7 |
0.7 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.7 |
2.9 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.7 |
2.9 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.7 |
3.6 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 |
2.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.7 |
4.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.7 |
2.6 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.7 |
2.6 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.6 |
12.7 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.6 |
3.2 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.6 |
23.5 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.6 |
0.6 |
GO:1902415 |
regulation of mRNA binding(GO:1902415) regulation of RNA binding(GO:1905214) |
| 0.6 |
3.1 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.6 |
23.1 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.6 |
0.6 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 0.6 |
1.8 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.6 |
0.6 |
GO:0042505 |
tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) |
| 0.6 |
2.4 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.6 |
0.6 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.6 |
1.8 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 |
1.8 |
GO:0001839 |
neural plate morphogenesis(GO:0001839) |
| 0.6 |
17.3 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.6 |
2.4 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.6 |
1.7 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.6 |
1.7 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.6 |
1.7 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.6 |
1.1 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.6 |
3.3 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.6 |
10.6 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.6 |
4.4 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.6 |
1.7 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 |
2.2 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 |
0.5 |
GO:0016078 |
tRNA catabolic process(GO:0016078) |
| 0.5 |
1.6 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.5 |
0.5 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.5 |
3.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.5 |
7.5 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.5 |
2.2 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 |
3.2 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 |
0.5 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.5 |
1.6 |
GO:0048382 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) mesendoderm development(GO:0048382) |
| 0.5 |
2.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.5 |
2.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.5 |
2.1 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.5 |
1.5 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) guanosine-containing compound catabolic process(GO:1901069) |
| 0.5 |
1.5 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.5 |
1.5 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.5 |
3.0 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.5 |
1.5 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.5 |
6.3 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.5 |
1.5 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.5 |
1.5 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.5 |
6.2 |
GO:0033120 |
positive regulation of RNA splicing(GO:0033120) |
| 0.5 |
1.4 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.5 |
1.9 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.5 |
0.9 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.5 |
8.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.5 |
5.1 |
GO:0070897 |
DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.5 |
1.9 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.5 |
3.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 |
3.2 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.5 |
1.4 |
GO:1904569 |
regulation of selenocysteine incorporation(GO:1904569) |
| 0.5 |
1.4 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.5 |
3.2 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.4 |
4.5 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 |
9.8 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.4 |
3.1 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.4 |
4.4 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 |
1.3 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 |
1.3 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.4 |
11.5 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.4 |
1.3 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 |
0.4 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 |
3.0 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 |
1.7 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.4 |
1.3 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.4 |
0.8 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.4 |
0.4 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.4 |
1.2 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.4 |
2.0 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.4 |
1.6 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.4 |
1.2 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.4 |
2.0 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.4 |
2.4 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.4 |
1.2 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.4 |
1.2 |
GO:0002681 |
somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 |
1.6 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.4 |
0.4 |
GO:0006404 |
RNA import into nucleus(GO:0006404) |
| 0.4 |
1.6 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.4 |
1.2 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 |
1.6 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 |
3.9 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 |
1.6 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 |
3.8 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.4 |
1.5 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 |
0.8 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 |
2.3 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 |
5.3 |
GO:0010388 |
cullin deneddylation(GO:0010388) |
| 0.4 |
2.2 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 |
1.1 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.4 |
5.6 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.4 |
2.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 |
0.7 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.4 |
2.2 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.4 |
7.5 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.4 |
2.1 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.4 |
1.1 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 |
1.1 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.4 |
1.8 |
GO:0060744 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.4 |
4.6 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.3 |
1.0 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.3 |
5.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.3 |
4.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.3 |
1.0 |
GO:0010986 |
complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.3 |
1.4 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.3 |
2.0 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.3 |
1.3 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.3 |
3.7 |
GO:1904851 |
positive regulation of establishment of protein localization to telomere(GO:1904851) |
| 0.3 |
1.0 |
GO:0042255 |
ribosome assembly(GO:0042255) |
| 0.3 |
1.7 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.3 |
5.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.3 |
2.0 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.3 |
1.6 |
GO:0090234 |
regulation of spindle assembly(GO:0090169) regulation of kinetochore assembly(GO:0090234) |
| 0.3 |
0.6 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.3 |
4.2 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 |
1.0 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.3 |
0.6 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 |
6.3 |
GO:0098534 |
centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.3 |
0.9 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
1.2 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 |
0.9 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.3 |
0.9 |
GO:0000722 |
telomere maintenance via recombination(GO:0000722) |
| 0.3 |
1.8 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.3 |
0.9 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 |
10.1 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.3 |
2.1 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.3 |
0.9 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.3 |
2.7 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.3 |
1.5 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 |
0.9 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.3 |
0.3 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 |
4.9 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.3 |
1.7 |
GO:0051096 |
positive regulation of helicase activity(GO:0051096) |
| 0.3 |
2.5 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.3 |
1.9 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.3 |
1.1 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
2.2 |
GO:0000963 |
mitochondrial RNA processing(GO:0000963) |
| 0.3 |
3.8 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.3 |
0.8 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 |
0.3 |
GO:0010824 |
regulation of centrosome duplication(GO:0010824) |
| 0.3 |
5.4 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.3 |
0.5 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.3 |
2.2 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.3 |
0.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.3 |
61.2 |
GO:0000398 |
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.3 |
1.1 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.3 |
2.4 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.3 |
1.3 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 |
4.7 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.3 |
0.5 |
GO:2000059 |
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.3 |
1.0 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.3 |
1.5 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.3 |
1.0 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 |
1.5 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 |
2.0 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.3 |
0.5 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 |
1.0 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 |
1.5 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.3 |
3.5 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.3 |
1.3 |
GO:0006167 |
AMP biosynthetic process(GO:0006167) |
| 0.3 |
0.8 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.3 |
1.0 |
GO:0045003 |
DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.3 |
1.8 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.3 |
0.8 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.3 |
0.3 |
GO:0002901 |
mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.2 |
2.2 |
GO:0006183 |
GTP biosynthetic process(GO:0006183) |
| 0.2 |
4.7 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.2 |
0.7 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 |
2.0 |
GO:0061085 |
regulation of histone H3-K27 methylation(GO:0061085) |
| 0.2 |
1.7 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.2 |
0.7 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.2 |
20.3 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.2 |
8.6 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.2 |
1.9 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.2 |
1.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.2 |
1.7 |
GO:0007320 |
insemination(GO:0007320) |
| 0.2 |
4.5 |
GO:0051654 |
establishment of mitochondrion localization(GO:0051654) |
| 0.2 |
0.7 |
GO:0045359 |
positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.2 |
1.2 |
GO:0018202 |
peptidyl-histidine modification(GO:0018202) |
| 0.2 |
2.1 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.2 |
0.9 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 |
0.7 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 |
0.2 |
GO:0036499 |
PERK-mediated unfolded protein response(GO:0036499) |
| 0.2 |
6.0 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.2 |
0.9 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.2 |
0.7 |
GO:0046078 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.2 |
0.4 |
GO:0006533 |
aspartate catabolic process(GO:0006533) |
| 0.2 |
2.9 |
GO:0032785 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.2 |
0.4 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.2 |
0.2 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.2 |
0.4 |
GO:0009838 |
abscission(GO:0009838) |
| 0.2 |
0.7 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 |
0.6 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.2 |
0.9 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.2 |
0.8 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 |
16.4 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.2 |
0.6 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.2 |
1.0 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.2 |
0.6 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
4.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.2 |
0.4 |
GO:0000087 |
mitotic M phase(GO:0000087) mitotic cell cycle phase(GO:0098763) |
| 0.2 |
0.2 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 |
0.2 |
GO:0060339 |
negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.2 |
0.4 |
GO:0046060 |
dATP metabolic process(GO:0046060) |
| 0.2 |
9.2 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.2 |
0.8 |
GO:0048659 |
smooth muscle cell proliferation(GO:0048659) |
| 0.2 |
1.4 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 |
1.0 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.2 |
0.8 |
GO:0085020 |
protein K6-linked ubiquitination(GO:0085020) |
| 0.2 |
1.2 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.2 |
0.8 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 |
0.8 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.2 |
3.8 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.2 |
1.3 |
GO:0032506 |
cytokinetic process(GO:0032506) |
| 0.2 |
0.6 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) chemoattraction of axon(GO:0061642) inner medullary collecting duct development(GO:0072061) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.2 |
0.2 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.2 |
0.2 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 |
0.4 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 |
0.6 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 |
0.4 |
GO:0010955 |
negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.2 |
0.2 |
GO:0045900 |
negative regulation of translational elongation(GO:0045900) |
| 0.2 |
1.1 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.2 |
1.8 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.2 |
0.4 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 |
0.7 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.2 |
1.1 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.2 |
1.1 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.2 |
0.7 |
GO:0090035 |
regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.2 |
0.4 |
GO:0070199 |
establishment of protein localization to chromosome(GO:0070199) |
| 0.2 |
2.1 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 |
1.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) |
| 0.2 |
1.8 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 |
0.5 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 |
0.9 |
GO:0032490 |
detection of molecule of bacterial origin(GO:0032490) |
| 0.2 |
0.9 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.2 |
0.3 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.2 |
1.9 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.2 |
0.7 |
GO:0061626 |
pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.2 |
2.7 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 |
0.7 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 |
2.5 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 |
0.7 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 |
0.2 |
GO:0030422 |
production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 |
1.3 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.2 |
1.3 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.6 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.2 |
1.4 |
GO:1901857 |
positive regulation of cellular respiration(GO:1901857) |
| 0.2 |
0.5 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 |
0.5 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 |
3.0 |
GO:0045589 |
regulation of regulatory T cell differentiation(GO:0045589) |
| 0.2 |
6.0 |
GO:0006418 |
tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 |
0.5 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.2 |
0.2 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.2 |
2.1 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 |
1.1 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.2 |
1.5 |
GO:0050942 |
positive regulation of developmental pigmentation(GO:0048087) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.2 |
1.7 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.2 |
1.1 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 |
0.6 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 |
0.4 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.7 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.1 |
0.6 |
GO:0070425 |
regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
3.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.3 |
GO:0035696 |
monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 |
2.2 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
1.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
1.3 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
4.4 |
GO:0000725 |
double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.1 |
1.5 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.7 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
15.8 |
GO:0008380 |
RNA splicing(GO:0008380) |
| 0.1 |
0.6 |
GO:0032784 |
regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 |
0.3 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 |
0.4 |
GO:0034969 |
histone arginine methylation(GO:0034969) |
| 0.1 |
1.2 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.1 |
0.4 |
GO:0008050 |
female courtship behavior(GO:0008050) |
| 0.1 |
1.0 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
0.7 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
1.6 |
GO:0007131 |
reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.1 |
0.6 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.1 |
1.4 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 |
0.3 |
GO:0070346 |
positive regulation of fat cell proliferation(GO:0070346) |
| 0.1 |
0.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
1.4 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.0 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
1.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 |
0.1 |
GO:1902237 |
positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.1 |
1.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
1.7 |
GO:0055070 |
copper ion homeostasis(GO:0055070) |
| 0.1 |
0.9 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.1 |
0.4 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 |
1.8 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 |
0.1 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.1 |
0.8 |
GO:0061009 |
common bile duct development(GO:0061009) |
| 0.1 |
0.3 |
GO:0006901 |
vesicle coating(GO:0006901) |
| 0.1 |
0.8 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.1 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
0.9 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
1.7 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 |
0.7 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 |
4.7 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 |
0.4 |
GO:0008334 |
histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.5 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 |
0.5 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
1.4 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
1.4 |
GO:0048535 |
lymph node development(GO:0048535) |
| 0.1 |
7.6 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 |
1.8 |
GO:0050000 |
chromosome localization(GO:0050000) establishment of chromosome localization(GO:0051303) |
| 0.1 |
0.5 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.1 |
1.0 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.1 |
0.2 |
GO:0045002 |
double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 |
2.6 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.1 |
0.6 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
1.6 |
GO:0071229 |
cellular response to acid chemical(GO:0071229) |
| 0.1 |
0.7 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 |
0.3 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 |
1.2 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.1 |
0.3 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
0.2 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
1.2 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.1 |
0.2 |
GO:0030656 |
regulation of vitamin metabolic process(GO:0030656) |
| 0.1 |
0.6 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 |
2.0 |
GO:1902236 |
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 |
0.9 |
GO:0036260 |
7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 |
0.8 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.1 |
1.1 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.1 |
0.4 |
GO:0046618 |
drug export(GO:0046618) |
| 0.1 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
1.3 |
GO:0009438 |
methylglyoxal metabolic process(GO:0009438) |
| 0.1 |
0.3 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.1 |
1.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
1.4 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
1.2 |
GO:0016126 |
sterol biosynthetic process(GO:0016126) |
| 0.1 |
1.2 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.1 |
1.3 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.1 |
0.3 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 |
0.7 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.1 |
0.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.1 |
0.8 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
1.2 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
0.6 |
GO:1904251 |
regulation of bile acid metabolic process(GO:1904251) |
| 0.1 |
0.4 |
GO:0036480 |
neuron intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0036480) regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903376) |
| 0.1 |
0.9 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.1 |
1.0 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.1 |
0.5 |
GO:0061101 |
neuroendocrine cell differentiation(GO:0061101) |
| 0.1 |
0.5 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.1 |
0.7 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.2 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.6 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
0.1 |
GO:0045066 |
CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) regulatory T cell differentiation(GO:0045066) |
| 0.1 |
2.4 |
GO:0010596 |
negative regulation of endothelial cell migration(GO:0010596) |
| 0.1 |
0.3 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.1 |
0.5 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.1 |
5.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.4 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.1 |
0.3 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 |
1.6 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
0.8 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.1 |
0.9 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 |
0.5 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.6 |
GO:1990564 |
protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
0.7 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
1.2 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 |
0.7 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 |
2.0 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.1 |
4.1 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.1 |
0.2 |
GO:0031397 |
negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 |
0.3 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 |
1.3 |
GO:0044819 |
mitotic G1/S transition checkpoint(GO:0044819) |
| 0.1 |
1.3 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.1 |
0.3 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 |
1.8 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.1 |
0.1 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 |
2.4 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.1 |
0.3 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.1 |
GO:1900368 |
RNA interference(GO:0016246) regulation of RNA interference(GO:1900368) |
| 0.1 |
6.5 |
GO:0042254 |
ribosome biogenesis(GO:0042254) |
| 0.1 |
0.9 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.1 |
0.6 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
6.0 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 |
0.3 |
GO:0052803 |
imidazole-containing compound metabolic process(GO:0052803) |
| 0.1 |
0.1 |
GO:1902953 |
positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
1.4 |
GO:0032786 |
positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 |
0.8 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
0.4 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 |
1.2 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.1 |
0.7 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
0.5 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 |
0.1 |
GO:1904429 |
t-circle formation(GO:0090656) regulation of t-circle formation(GO:1904429) |
| 0.1 |
0.2 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.7 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.1 |
2.2 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
2.7 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.1 |
0.4 |
GO:0039532 |
negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 |
0.2 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.4 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
1.3 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 |
0.5 |
GO:0006699 |
bile acid biosynthetic process(GO:0006699) |
| 0.1 |
2.5 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 |
0.7 |
GO:0009065 |
glutamine family amino acid catabolic process(GO:0009065) |
| 0.1 |
0.2 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
0.8 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 |
0.3 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.2 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
0.2 |
GO:0035928 |
RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) |
| 0.1 |
0.8 |
GO:0015825 |
L-serine transport(GO:0015825) |
| 0.1 |
0.8 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.7 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
0.2 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.1 |
2.2 |
GO:2000147 |
positive regulation of cell motility(GO:2000147) |
| 0.1 |
0.8 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.4 |
GO:0038030 |
non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 |
0.2 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 |
0.3 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
0.2 |
GO:0006702 |
androgen biosynthetic process(GO:0006702) |
| 0.1 |
0.3 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.1 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 |
0.2 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.1 |
1.4 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.1 |
0.6 |
GO:0090043 |
tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.1 |
0.4 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.1 |
0.2 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 |
0.2 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
0.7 |
GO:0035308 |
negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 |
0.3 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 |
0.7 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.4 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.9 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.1 |
0.3 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
0.3 |
GO:0051531 |
NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 |
0.7 |
GO:0060013 |
righting reflex(GO:0060013) |
| 0.1 |
0.1 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.1 |
1.4 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 |
0.6 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.3 |
GO:0001824 |
blastocyst development(GO:0001824) |
| 0.1 |
1.4 |
GO:0000578 |
embryonic axis specification(GO:0000578) |
| 0.1 |
2.0 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.1 |
0.1 |
GO:1902177 |
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 |
1.5 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.1 |
GO:0007227 |
signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.7 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 |
0.2 |
GO:0014891 |
striated muscle atrophy(GO:0014891) |
| 0.1 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
2.7 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.1 |
0.5 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.2 |
GO:0043323 |
natural killer cell degranulation(GO:0043320) regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
2.2 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 |
1.4 |
GO:0071482 |
cellular response to light stimulus(GO:0071482) |
| 0.1 |
0.3 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
0.3 |
GO:1901341 |
activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 |
0.5 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
1.8 |
GO:0031016 |
pancreas development(GO:0031016) |
| 0.1 |
0.2 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.1 |
0.4 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 |
2.1 |
GO:0035307 |
positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 |
2.3 |
GO:0031570 |
DNA integrity checkpoint(GO:0031570) |
| 0.1 |
0.2 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
0.3 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 |
0.2 |
GO:0045324 |
late endosome to vacuole transport(GO:0045324) |
| 0.1 |
0.3 |
GO:0070243 |
regulation of thymocyte apoptotic process(GO:0070243) |
| 0.1 |
0.9 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 |
0.2 |
GO:1901301 |
regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 |
2.8 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.0 |
0.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.5 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.8 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.1 |
GO:0045896 |
regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 |
0.7 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.6 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.8 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
1.4 |
GO:0007368 |
determination of left/right symmetry(GO:0007368) |
| 0.0 |
0.2 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
2.5 |
GO:0042476 |
odontogenesis(GO:0042476) |
| 0.0 |
0.1 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 |
0.3 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
1.0 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.5 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 |
0.3 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.1 |
GO:0042510 |
tyrosine phosphorylation of Stat1 protein(GO:0042508) regulation of tyrosine phosphorylation of Stat1 protein(GO:0042510) positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.0 |
1.2 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.2 |
GO:1904469 |
positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 |
0.9 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.3 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.0 |
0.2 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.8 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.1 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.4 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 |
0.2 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.3 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0003273 |
cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 |
1.0 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
1.0 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.0 |
0.2 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.3 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
1.8 |
GO:0010950 |
positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 |
0.1 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 |
2.0 |
GO:0009952 |
anterior/posterior pattern specification(GO:0009952) |
| 0.0 |
0.1 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 |
0.5 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.2 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.4 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.3 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.1 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.5 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.3 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.1 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.0 |
1.8 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.0 |
0.2 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.1 |
GO:0010717 |
regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 |
0.4 |
GO:1903427 |
negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 |
0.2 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.3 |
GO:0097049 |
motor neuron apoptotic process(GO:0097049) regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
5.7 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.2 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.0 |
0.1 |
GO:1990253 |
cellular response to leucine starvation(GO:1990253) |
| 0.0 |
1.0 |
GO:0071346 |
cellular response to interferon-gamma(GO:0071346) |
| 0.0 |
0.3 |
GO:1900025 |
negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 |
0.6 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.6 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 |
0.1 |
GO:0019255 |
glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 |
0.2 |
GO:0002484 |
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 |
0.8 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 |
0.7 |
GO:2001243 |
negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 |
0.7 |
GO:0042267 |
natural killer cell mediated immunity(GO:0002228) natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 |
0.7 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.0 |
GO:0061051 |
positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 |
0.8 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.2 |
GO:0060068 |
female genitalia development(GO:0030540) vagina development(GO:0060068) |
| 0.0 |
6.2 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
1.1 |
GO:0050868 |
negative regulation of T cell activation(GO:0050868) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
1.2 |
GO:0048008 |
platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 |
0.1 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.8 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.0 |
0.2 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.7 |
GO:0006633 |
fatty acid biosynthetic process(GO:0006633) |
| 0.0 |
0.6 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:0060948 |
cardiac endothelial cell differentiation(GO:0003348) epicardium-derived cardiac endothelial cell differentiation(GO:0003349) cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 |
0.3 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.2 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.4 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.1 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.3 |
GO:0045930 |
negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.5 |
GO:1902593 |
protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 |
0.2 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0006743 |
ubiquinone metabolic process(GO:0006743) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.2 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
4.1 |
GO:0032259 |
methylation(GO:0032259) |
| 0.0 |
0.7 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.1 |
GO:0015936 |
coenzyme A metabolic process(GO:0015936) |
| 0.0 |
0.3 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.0 |
0.6 |
GO:0045669 |
positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 |
0.3 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 |
5.2 |
GO:0006412 |
translation(GO:0006412) |
| 0.0 |
0.6 |
GO:0006626 |
protein targeting to mitochondrion(GO:0006626) |
| 0.0 |
0.1 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.0 |
0.1 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.4 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.2 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
0.1 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.3 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.1 |
GO:0006788 |
heme oxidation(GO:0006788) |
| 0.0 |
0.5 |
GO:0050770 |
regulation of axonogenesis(GO:0050770) |
| 0.0 |
0.6 |
GO:0050731 |
positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 |
0.4 |
GO:0090181 |
regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 |
0.1 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.3 |
GO:0051193 |
regulation of cofactor metabolic process(GO:0051193) |
| 0.0 |
0.3 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.1 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.5 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.0 |
0.1 |
GO:0033866 |
coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 |
0.1 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.0 |
0.0 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.1 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.0 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.4 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.0 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.1 |
GO:0051791 |
medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 |
0.0 |
GO:0010726 |
regulation of hydrogen peroxide metabolic process(GO:0010310) positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.0 |
GO:0001837 |
epithelial to mesenchymal transition(GO:0001837) |
| 0.0 |
0.2 |
GO:0001890 |
placenta development(GO:0001890) |
| 0.0 |
0.0 |
GO:0035358 |
regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035358) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.0 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 |
0.1 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |