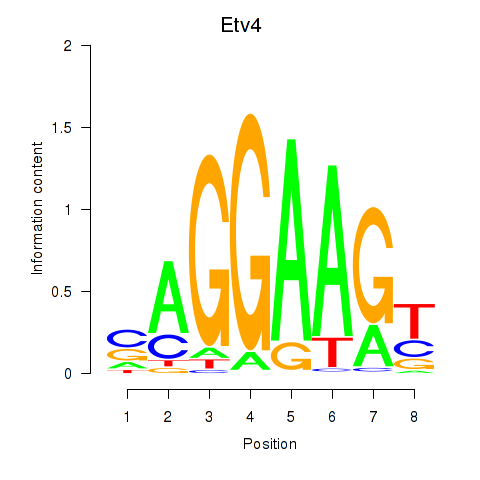
| 0.9 |
7.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.7 |
2.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.6 |
1.9 |
GO:0019043 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) |
| 0.6 |
2.5 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.6 |
1.7 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.5 |
1.6 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.5 |
2.4 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.5 |
1.9 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.5 |
2.3 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.4 |
1.8 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.4 |
4.4 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) |
| 0.4 |
0.4 |
GO:2000277 |
positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.4 |
0.4 |
GO:0002432 |
granuloma formation(GO:0002432) |
| 0.4 |
1.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.4 |
1.8 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.4 |
1.4 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.3 |
1.4 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.3 |
1.6 |
GO:0019086 |
late viral transcription(GO:0019086) |
| 0.3 |
1.6 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.3 |
1.9 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.3 |
1.8 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 |
1.7 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.3 |
1.4 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.3 |
3.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 |
0.8 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 |
2.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
1.5 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.2 |
1.4 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 |
0.7 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 |
1.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
0.7 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 |
0.9 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 |
1.8 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.2 |
0.7 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 |
2.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
0.9 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.2 |
0.6 |
GO:2001187 |
positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.2 |
3.3 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 |
0.8 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 |
0.6 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 |
0.8 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 |
1.2 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.2 |
1.2 |
GO:0070278 |
extracellular matrix constituent secretion(GO:0070278) |
| 0.2 |
1.4 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
1.0 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.2 |
0.4 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.2 |
1.8 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.2 |
0.5 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 |
0.4 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 |
1.1 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 |
0.9 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 |
0.5 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.2 |
1.5 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
1.9 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.2 |
1.2 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 |
2.2 |
GO:0010447 |
response to acidic pH(GO:0010447) |
| 0.2 |
2.8 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
0.6 |
GO:0021886 |
hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 |
0.6 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 |
0.8 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
1.2 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 |
0.4 |
GO:0014042 |
regulation of mitochondrial fusion(GO:0010635) positive regulation of mitochondrial fusion(GO:0010636) positive regulation of neuron maturation(GO:0014042) |
| 0.1 |
1.3 |
GO:0090045 |
positive regulation of deacetylase activity(GO:0090045) |
| 0.1 |
0.4 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 |
1.0 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.1 |
0.9 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.1 |
0.4 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.7 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 |
5.9 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 |
0.4 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
0.4 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 |
0.5 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
0.9 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.1 |
0.7 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) |
| 0.1 |
0.2 |
GO:0032661 |
regulation of interleukin-18 production(GO:0032661) negative regulation of interleukin-18 production(GO:0032701) |
| 0.1 |
1.0 |
GO:0090166 |
Golgi disassembly(GO:0090166) |
| 0.1 |
0.5 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.5 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 |
0.8 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.1 |
0.9 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 |
0.3 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.1 |
0.7 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.1 |
0.8 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 |
0.3 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.7 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
0.7 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 |
2.0 |
GO:0007095 |
mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 |
1.2 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 |
0.3 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.3 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
0.7 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 |
0.6 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.7 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
1.2 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
1.3 |
GO:2000009 |
negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 |
0.7 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.3 |
GO:0050859 |
negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 |
0.5 |
GO:0015871 |
choline transport(GO:0015871) regulation of resting membrane potential(GO:0060075) |
| 0.1 |
0.5 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.1 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.3 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
2.2 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.1 |
4.2 |
GO:0047496 |
vesicle transport along microtubule(GO:0047496) |
| 0.1 |
1.1 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.1 |
0.4 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
0.3 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 |
0.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.3 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) sensory perception of bitter taste(GO:0050913) |
| 0.1 |
0.4 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
0.2 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.3 |
GO:1900110 |
negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.1 |
0.2 |
GO:0002679 |
respiratory burst involved in defense response(GO:0002679) neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.3 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.9 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.5 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.1 |
0.3 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.1 |
0.4 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 |
2.0 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 |
0.5 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 |
1.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.3 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
0.3 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.5 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 |
0.3 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 |
1.1 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
2.8 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.1 |
0.4 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.3 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.7 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
1.2 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.1 |
0.5 |
GO:0008334 |
histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.2 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 |
0.5 |
GO:1902285 |
semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 |
0.5 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
1.8 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.1 |
1.7 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
0.3 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.2 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 |
0.2 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.9 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
0.3 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.1 |
0.5 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
0.2 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.3 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.1 |
0.5 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.1 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.1 |
0.4 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.0 |
0.2 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.0 |
0.2 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 |
0.5 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 |
1.6 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.8 |
GO:0042554 |
superoxide anion generation(GO:0042554) |
| 0.0 |
0.7 |
GO:0016559 |
peroxisome fission(GO:0016559) |
| 0.0 |
3.0 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.4 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
1.2 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.5 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.4 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
3.1 |
GO:0060976 |
coronary vasculature development(GO:0060976) |
| 0.0 |
0.3 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.8 |
GO:1900078 |
positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 |
0.2 |
GO:0010988 |
regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 |
0.1 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 |
0.4 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.2 |
GO:0090085 |
regulation of protein deubiquitination(GO:0090085) |
| 0.0 |
0.3 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 |
0.1 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.0 |
0.4 |
GO:0048741 |
myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.3 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.5 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.2 |
GO:0033632 |
regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 |
5.4 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.1 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.0 |
0.3 |
GO:0010759 |
positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 |
0.2 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
0.9 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
1.2 |
GO:0008631 |
intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 |
0.6 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.5 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.6 |
GO:0051352 |
negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) |
| 0.0 |
0.1 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 |
0.2 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 |
0.5 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 |
0.4 |
GO:0044144 |
regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 |
0.2 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
1.3 |
GO:0071346 |
cellular response to interferon-gamma(GO:0071346) |
| 0.0 |
0.4 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.5 |
GO:0030204 |
chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 |
0.7 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 |
0.3 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.3 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 |
0.4 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 |
2.0 |
GO:1901379 |
regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 |
1.3 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |
| 0.0 |
1.6 |
GO:0007585 |
respiratory gaseous exchange(GO:0007585) |
| 0.0 |
1.2 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 |
0.3 |
GO:0048711 |
positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 |
0.1 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.3 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.2 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 |
0.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.8 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
0.5 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
1.6 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.1 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.7 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.0 |
0.9 |
GO:0035162 |
embryonic hemopoiesis(GO:0035162) |
| 0.0 |
0.2 |
GO:0002755 |
MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 |
0.3 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.0 |
1.0 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
1.4 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.1 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0046929 |
negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
1.1 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.7 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.1 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.0 |
0.2 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 |
0.6 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.7 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.6 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.8 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
2.5 |
GO:0010977 |
negative regulation of neuron projection development(GO:0010977) |
| 0.0 |
0.4 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.2 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.5 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.9 |
GO:0032543 |
mitochondrial translation(GO:0032543) |
| 0.0 |
0.4 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.3 |
GO:0000737 |
DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 |
0.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.0 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 |
0.3 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.1 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.9 |
GO:0045665 |
negative regulation of neuron differentiation(GO:0045665) |
| 0.0 |
0.3 |
GO:0098927 |
early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.6 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.1 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.2 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.0 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.3 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.0 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 |
0.2 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 |
0.1 |
GO:0007096 |
regulation of exit from mitosis(GO:0007096) |
| 0.0 |
0.2 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |