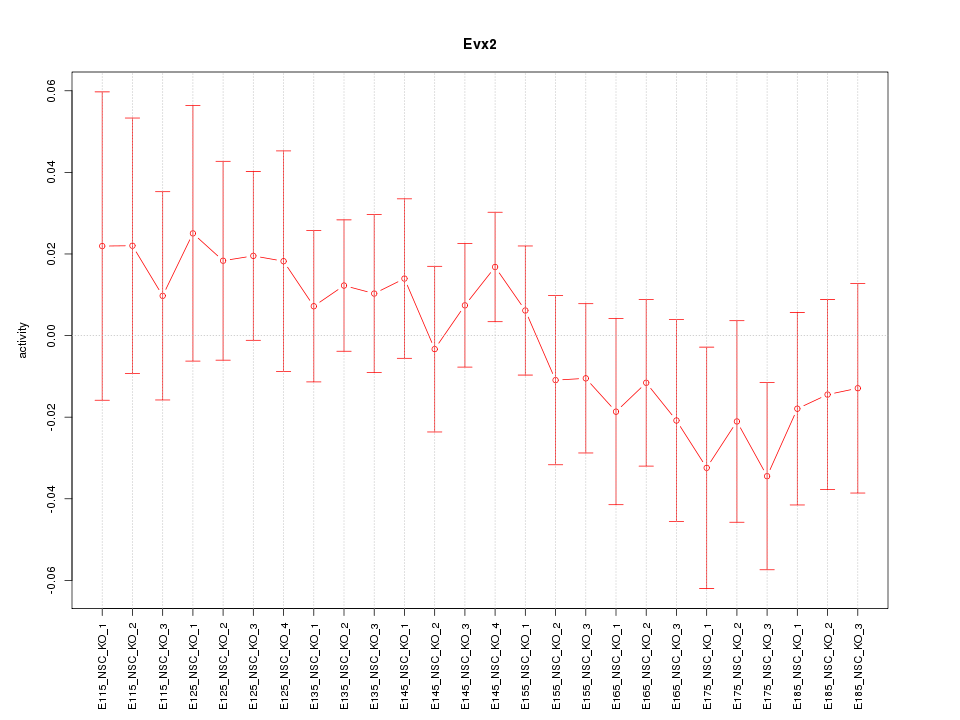
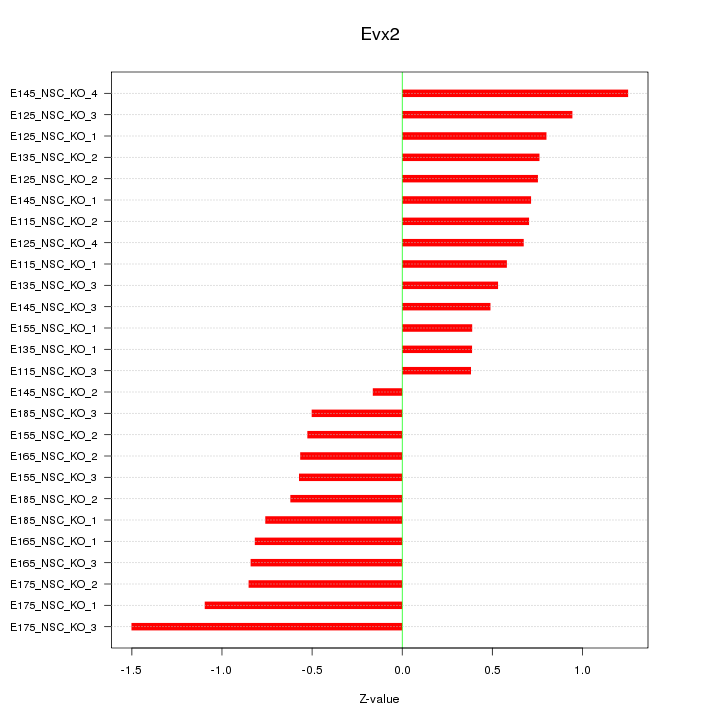
Motif ID: Evx2
Z-value: 0.753

Transcription factors associated with Evx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Evx2 | ENSMUSG00000001815.9 | Evx2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.5 | 1.4 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.4 | 7.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 0.9 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 3.5 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 0.7 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.2 | 0.9 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 1.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 2.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.8 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.3 | GO:0046469 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.2 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 3.0 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 2.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.5 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 9.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 2.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 2.0 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 12.4 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |