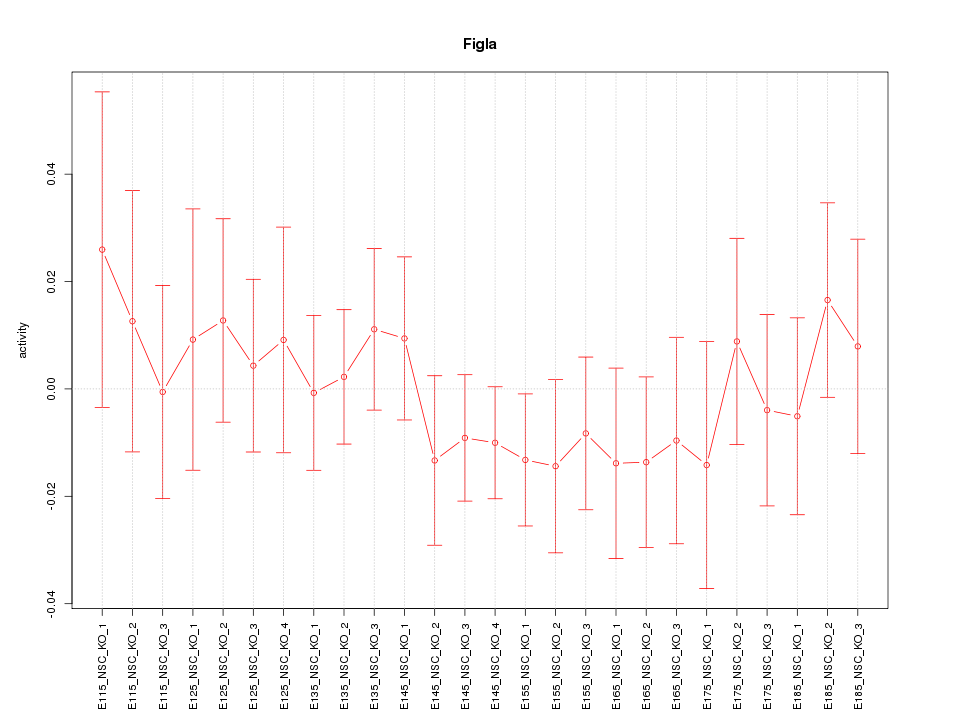
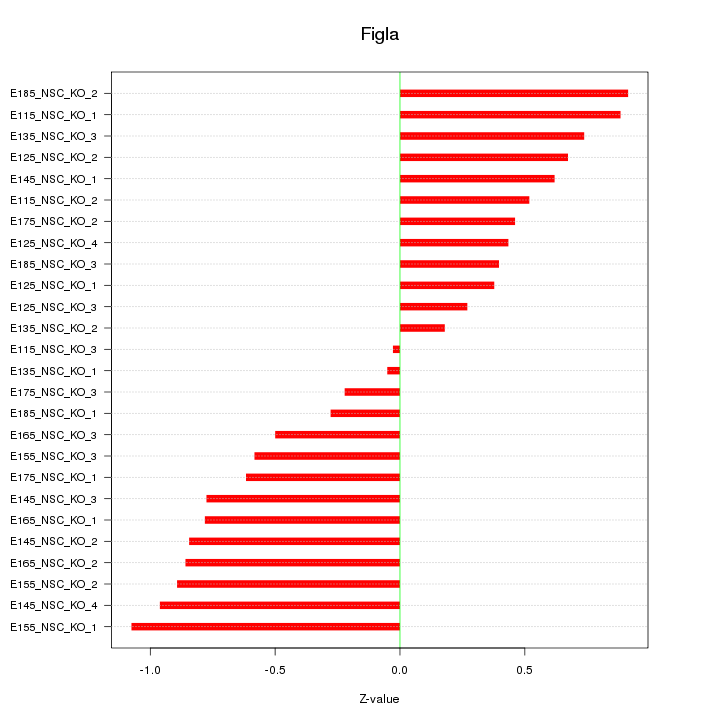
| 1.0 |
3.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 1.0 |
3.1 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.7 |
2.8 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.3 |
2.3 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.3 |
1.0 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.3 |
1.2 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 |
0.9 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 |
1.2 |
GO:0051311 |
spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.3 |
1.4 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.3 |
1.1 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.3 |
0.8 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 |
1.0 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.2 |
1.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
1.0 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.2 |
0.9 |
GO:0043987 |
histone H3-S10 phosphorylation(GO:0043987) |
| 0.2 |
0.7 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.2 |
0.6 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 |
1.1 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 |
0.4 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.2 |
0.8 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.2 |
0.8 |
GO:0003223 |
ventricular compact myocardium morphogenesis(GO:0003223) proprioception(GO:0019230) |
| 0.2 |
0.8 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
1.8 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
0.4 |
GO:0006116 |
NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 |
0.5 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 |
0.4 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 |
0.4 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.5 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.1 |
0.8 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.6 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.1 |
3.8 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
1.0 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.9 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.1 |
0.4 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.4 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.2 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
0.3 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.8 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.8 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.1 |
0.5 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
0.8 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.7 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.1 |
0.5 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.1 |
0.4 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
0.3 |
GO:1990379 |
lipid transport across blood brain barrier(GO:1990379) |
| 0.1 |
0.4 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.1 |
0.3 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.4 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
2.2 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.9 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.1 |
0.3 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 |
0.5 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.3 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.5 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.0 |
0.3 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.4 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.2 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 |
0.7 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
1.3 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.1 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 |
0.2 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.4 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 |
0.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.0 |
0.1 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 |
0.5 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.1 |
GO:0098908 |
regulation of neuronal action potential(GO:0098908) |
| 0.0 |
0.3 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.6 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 |
0.5 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.3 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
1.6 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.3 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.2 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.6 |
GO:0001523 |
retinoid metabolic process(GO:0001523) |
| 0.0 |
0.1 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.0 |
0.3 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.2 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.6 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 |
0.1 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.4 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.4 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.7 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.5 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.2 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.1 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.0 |
1.4 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.4 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.1 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.0 |
0.4 |
GO:0006739 |
NADP metabolic process(GO:0006739) |