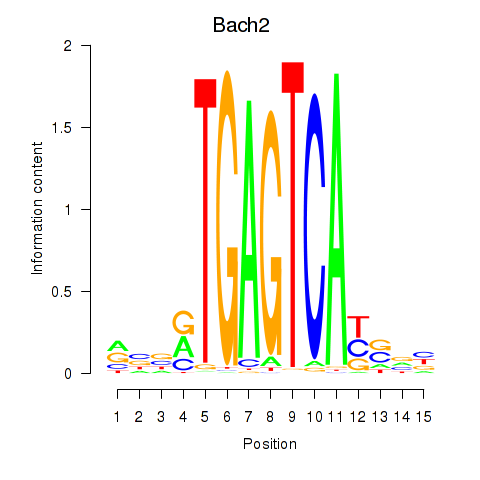
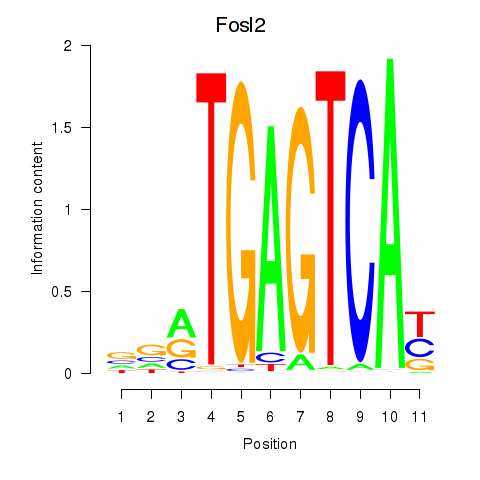
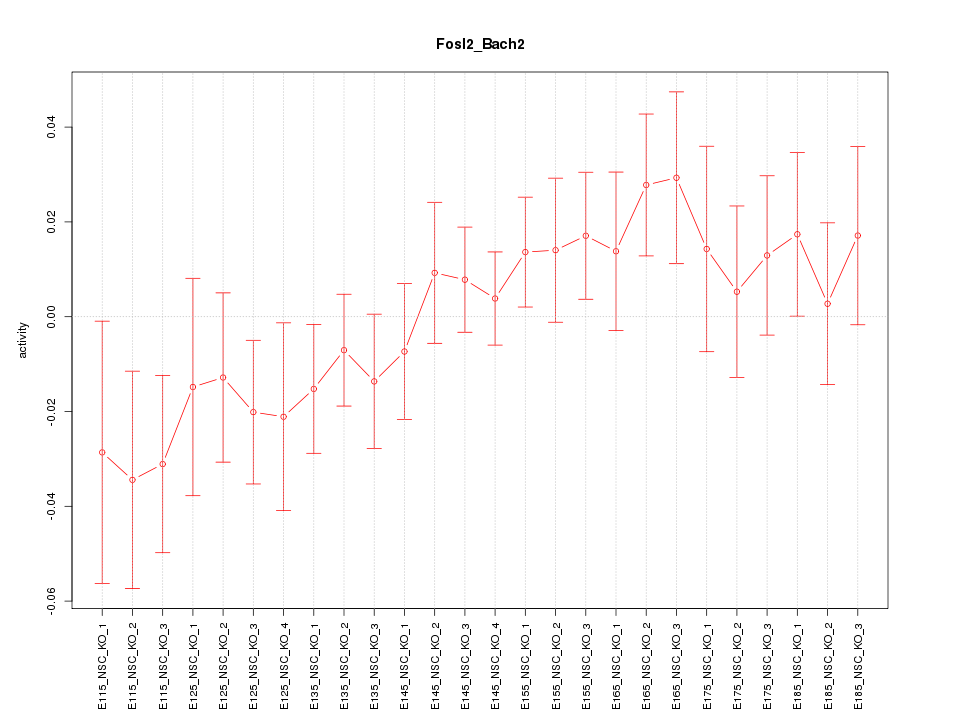
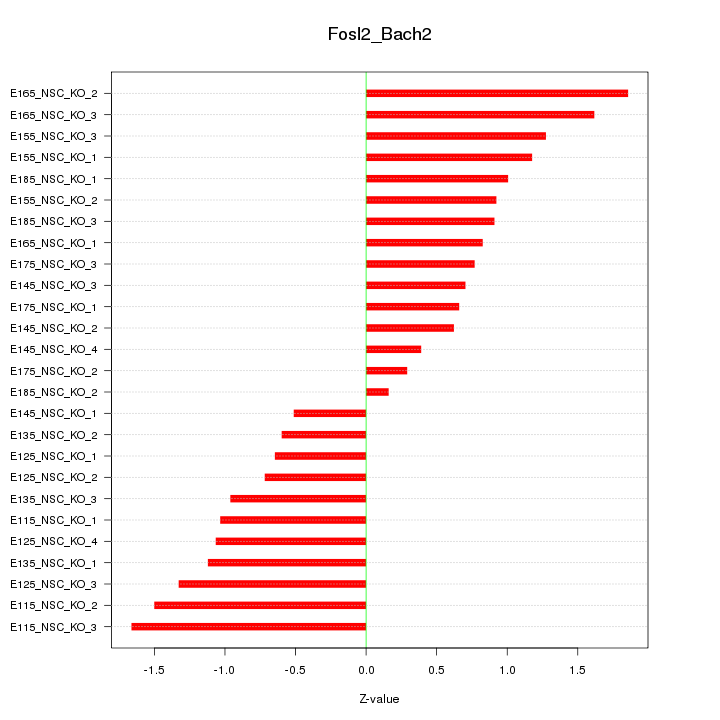
| 2.1 |
8.6 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 1.6 |
6.5 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.6 |
6.3 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 1.5 |
4.5 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 1.3 |
8.0 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 1.1 |
4.5 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 1.0 |
8.8 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.9 |
2.8 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.7 |
4.5 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 |
2.9 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.7 |
6.5 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 |
3.8 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.6 |
3.7 |
GO:0046103 |
ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.6 |
4.9 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.6 |
2.2 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.5 |
3.2 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 |
1.5 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.5 |
4.5 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.5 |
1.9 |
GO:0070885 |
negative regulation of interleukin-17 production(GO:0032700) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.5 |
3.4 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.5 |
1.4 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.5 |
3.7 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.4 |
1.7 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.4 |
1.2 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 |
1.5 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.4 |
1.8 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.4 |
1.1 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.4 |
1.4 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.4 |
1.1 |
GO:0051464 |
positive regulation of cortisol secretion(GO:0051464) |
| 0.4 |
3.9 |
GO:2001135 |
regulation of endocytic recycling(GO:2001135) |
| 0.3 |
2.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.3 |
3.5 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.3 |
1.0 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 |
0.9 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 |
1.2 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.3 |
0.3 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.3 |
1.1 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.3 |
1.1 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.3 |
10.2 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 |
3.6 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.3 |
0.8 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 |
1.3 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) phospholipid efflux(GO:0033700) |
| 0.2 |
0.7 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 |
1.7 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.2 |
0.8 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.2 |
0.6 |
GO:0090272 |
negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.2 |
2.0 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.2 |
0.9 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.2 |
0.9 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.2 |
2.9 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.2 |
3.7 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.2 |
1.1 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.2 |
2.2 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 |
1.8 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.2 |
0.8 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.9 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
9.8 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
1.3 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
0.4 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
4.9 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 |
0.8 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.4 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
2.4 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 |
0.9 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 |
0.4 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.8 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.5 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
0.8 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.2 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 |
1.4 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
1.0 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
2.1 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.1 |
0.8 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 |
0.5 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
1.5 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
0.4 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.1 |
0.4 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
1.6 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.4 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.1 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 |
2.5 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.7 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.3 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 |
1.4 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
3.4 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 |
1.6 |
GO:0034113 |
heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 |
2.3 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.1 |
2.6 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.7 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 |
2.8 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 |
0.5 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
0.7 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 |
0.4 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.5 |
GO:0097576 |
autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.1 |
0.7 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.1 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.5 |
GO:0090129 |
positive regulation of synapse maturation(GO:0090129) |
| 0.1 |
0.7 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 |
1.7 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
0.5 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.6 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 |
1.3 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.1 |
1.3 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
1.4 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
0.7 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.2 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
1.8 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.0 |
3.5 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.4 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.2 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.4 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
3.0 |
GO:0042475 |
odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 |
0.7 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.4 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.9 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.6 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.5 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.6 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
2.5 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.3 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 |
0.2 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.4 |
GO:0044342 |
type B pancreatic cell proliferation(GO:0044342) |
| 0.0 |
1.0 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
1.0 |
GO:0051438 |
regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 |
0.6 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.0 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.0 |
2.7 |
GO:0006275 |
regulation of DNA replication(GO:0006275) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.5 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.2 |
GO:0071451 |
cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 |
0.1 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.0 |
0.1 |
GO:0016584 |
nucleosome positioning(GO:0016584) |