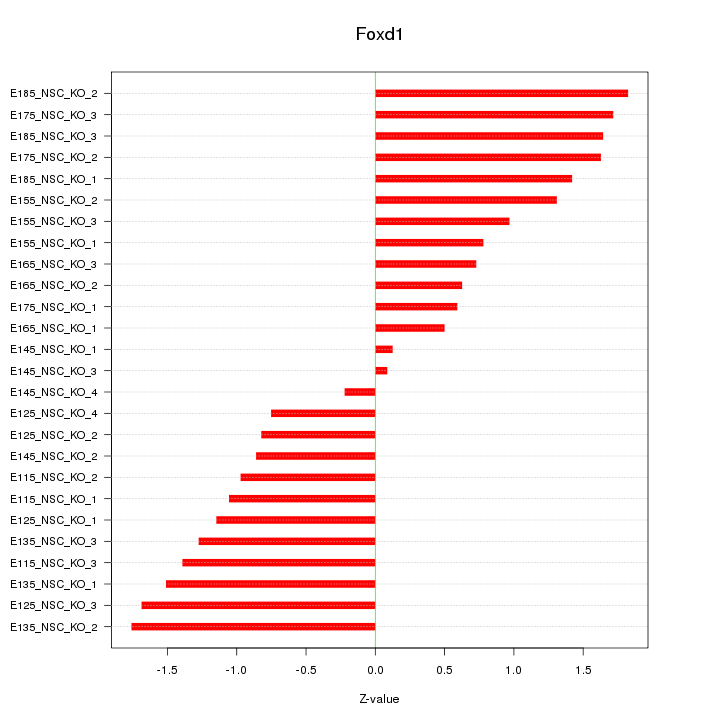
Motif ID: Foxd1
Z-value: 1.171
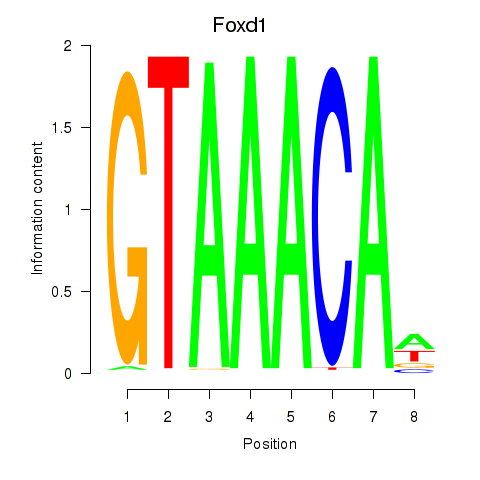
Transcription factors associated with Foxd1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxd1 | ENSMUSG00000078302.3 | Foxd1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxd1 | mm10_v2_chr13_+_98354234_98354250 | -0.20 | 3.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 19.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.6 | 4.7 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 1.4 | 4.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.4 | 9.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.4 | 4.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.3 | 6.4 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.2 | 4.8 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.1 | 4.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.9 | 2.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.8 | 2.5 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.8 | 2.3 | GO:1901254 | positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.6 | 1.9 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.6 | 1.9 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.6 | 4.3 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.6 | 2.8 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.6 | 2.8 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.6 | 1.7 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 | 7.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.5 | 2.0 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 1.9 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.5 | 2.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.5 | 3.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.4 | 4.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 | 1.3 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.4 | 2.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.4 | 7.2 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.4 | 3.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.4 | 4.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 3.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.4 | 1.2 | GO:0003032 | detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.4 | 1.2 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.4 | 2.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.4 | 1.5 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.3 | 3.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 2.1 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.3 | 1.7 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.3 | 2.7 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 4.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.3 | 1.3 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.3 | 1.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 1.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.2 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 4.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 2.3 | GO:0098532 | liver regeneration(GO:0097421) histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 0.7 | GO:0071336 | lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 | 0.7 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 1.4 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.2 | 0.6 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.2 | 2.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 | 2.5 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 1.6 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 2.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 3.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.2 | 3.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 1.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.1 | 1.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 2.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.1 | 1.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.9 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 1.6 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.9 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 2.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.9 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 1.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 0.9 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.7 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.5 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 1.7 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 1.6 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.8 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.4 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.2 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 | 0.4 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 1.8 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.9 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.3 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.4 | GO:0043970 | histone H3-K9 acetylation(GO:0043970) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 4.8 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 1.5 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.8 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.7 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0042025 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.7 | 3.0 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 6.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.4 | 1.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 2.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 4.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.7 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.8 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 2.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 4.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 4.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 2.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 2.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.2 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 1.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 1.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 12.9 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 4.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 7.2 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 5.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 5.2 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.0 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 4.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 2.8 | 19.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.4 | 4.2 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 1.0 | 4.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 2.6 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.7 | 2.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.6 | 1.9 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.6 | 2.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.6 | 1.9 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.6 | 2.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.5 | 5.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 2.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.5 | 1.9 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 3.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.5 | 2.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 1.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 4.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.4 | 2.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 1.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 4.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.3 | 1.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 0.9 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 2.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 1.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 2.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.9 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 3.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.7 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.2 | 1.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 3.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.5 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 2.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.5 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 3.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.6 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.1 | 0.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 3.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.0 | 4.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 3.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.7 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 0.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 1.3 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 2.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |