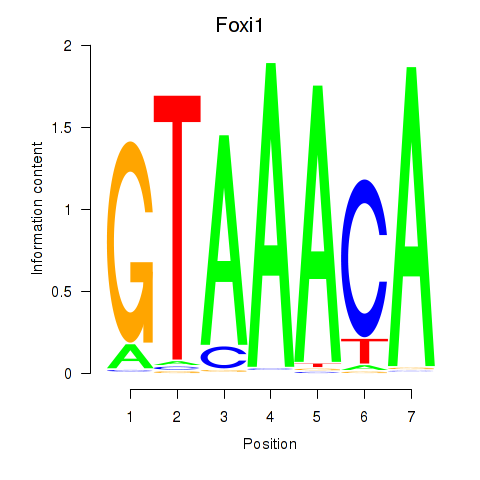
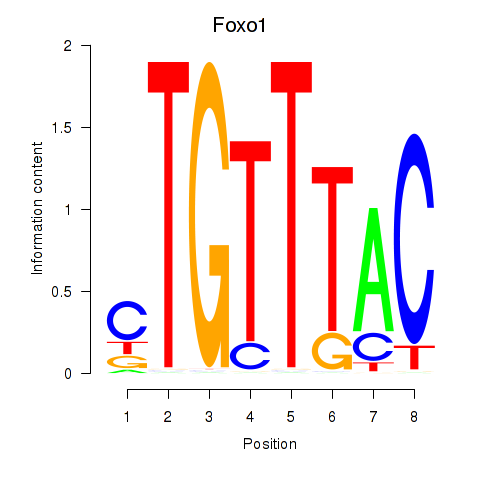
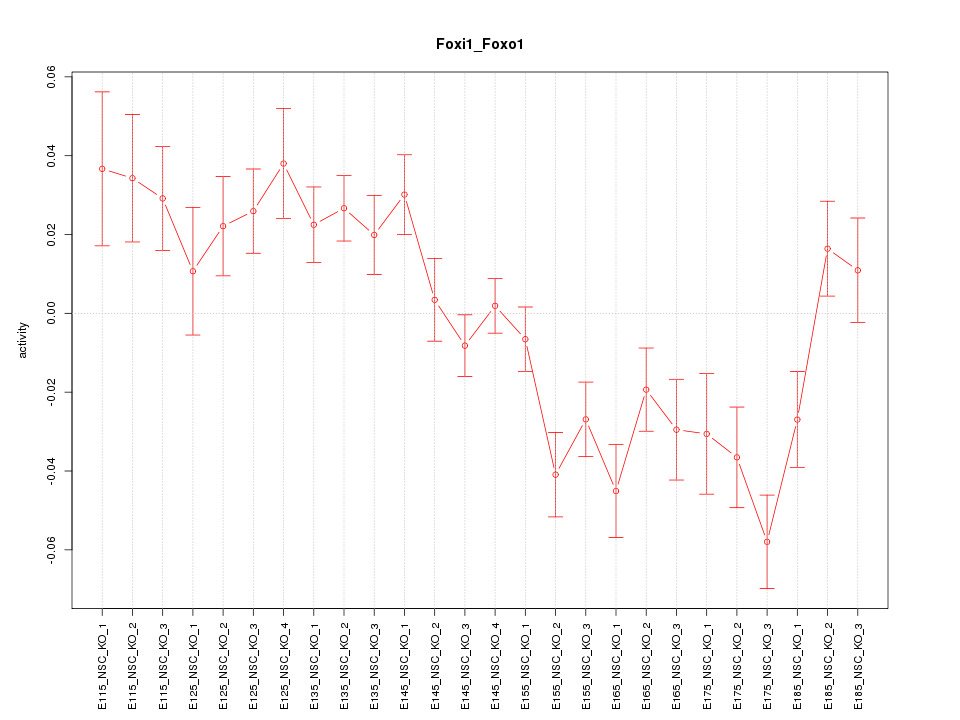
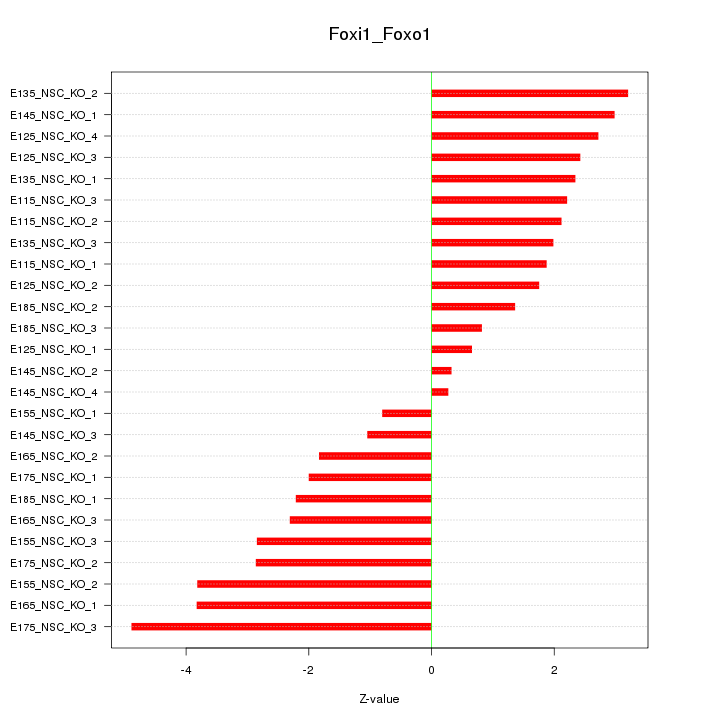
| 4.9 |
29.7 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 3.5 |
17.3 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 3.3 |
9.8 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 3.2 |
9.7 |
GO:0021557 |
oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 3.0 |
9.1 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 2.7 |
2.7 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 2.6 |
7.7 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 2.4 |
21.4 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 2.2 |
11.2 |
GO:0031052 |
programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 2.2 |
8.6 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 2.1 |
6.2 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.0 |
5.9 |
GO:0042908 |
xenobiotic transport(GO:0042908) |
| 1.6 |
4.7 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.6 |
28.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.5 |
28.7 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 1.5 |
7.5 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.5 |
8.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.4 |
17.7 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 1.4 |
14.9 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 1.4 |
9.5 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 1.3 |
2.7 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 1.3 |
3.9 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 1.3 |
3.8 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 1.2 |
4.9 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 1.2 |
13.4 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 1.2 |
4.8 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 1.2 |
3.6 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 1.2 |
3.5 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 1.1 |
12.5 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 1.1 |
3.4 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 1.1 |
4.5 |
GO:2000675 |
negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 1.1 |
3.3 |
GO:0002339 |
B cell selection(GO:0002339) |
| 1.1 |
9.9 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 1.1 |
6.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 1.1 |
3.3 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 1.1 |
20.6 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 1.1 |
4.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 1.1 |
2.1 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 1.0 |
5.2 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.0 |
4.2 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 1.0 |
22.1 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 1.0 |
3.0 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 1.0 |
1.0 |
GO:0097694 |
establishment of RNA localization to telomere(GO:0097694) |
| 1.0 |
12.8 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 1.0 |
5.8 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 1.0 |
3.9 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.9 |
2.8 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.9 |
4.7 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.9 |
2.8 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.9 |
8.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.9 |
1.8 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.9 |
8.0 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.9 |
3.5 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.9 |
2.6 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.9 |
4.3 |
GO:2000668 |
dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.9 |
4.3 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.9 |
4.3 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.8 |
1.7 |
GO:0060528 |
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.8 |
1.7 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.8 |
4.2 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.8 |
6.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.8 |
4.1 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.8 |
4.1 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.8 |
4.1 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.8 |
7.1 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.8 |
2.3 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.8 |
2.3 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.8 |
2.3 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.7 |
2.9 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.7 |
8.0 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.7 |
7.8 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.7 |
4.2 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.7 |
1.4 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.7 |
2.0 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.7 |
1.3 |
GO:0002765 |
immune response-inhibiting signal transduction(GO:0002765) |
| 0.7 |
3.3 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.7 |
2.0 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.7 |
2.7 |
GO:0060287 |
epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.7 |
0.7 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.6 |
3.2 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.6 |
1.9 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.6 |
1.9 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.6 |
3.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.6 |
3.7 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.6 |
1.8 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.6 |
3.1 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.6 |
1.8 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 |
2.4 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.6 |
11.9 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.6 |
2.3 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.6 |
0.6 |
GO:0010579 |
regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.6 |
8.0 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.6 |
5.7 |
GO:0060179 |
male mating behavior(GO:0060179) |
| 0.6 |
1.7 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.6 |
7.3 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.6 |
2.2 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.6 |
2.8 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.6 |
1.7 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.6 |
3.9 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.6 |
0.6 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.6 |
1.7 |
GO:1902071 |
regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.5 |
2.7 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.5 |
10.3 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.5 |
4.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.5 |
1.1 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.5 |
2.7 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.5 |
1.6 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 |
10.7 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.5 |
5.8 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.5 |
3.1 |
GO:1903441 |
protein localization to ciliary membrane(GO:1903441) |
| 0.5 |
2.6 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.5 |
1.0 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.5 |
2.1 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.5 |
2.0 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.5 |
2.6 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 |
1.5 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.5 |
1.5 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.5 |
1.5 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.5 |
2.0 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) |
| 0.5 |
2.5 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.5 |
1.5 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.5 |
5.5 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.5 |
4.5 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.5 |
2.0 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.5 |
1.5 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.5 |
1.5 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.5 |
1.9 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.5 |
1.0 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.5 |
4.7 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.5 |
1.9 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.5 |
14.1 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.5 |
2.8 |
GO:0051096 |
telomere assembly(GO:0032202) regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.5 |
1.4 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.5 |
1.8 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.5 |
1.4 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.5 |
1.8 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.5 |
1.8 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.4 |
2.2 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.4 |
1.3 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.4 |
2.7 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.4 |
1.3 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.4 |
2.2 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.4 |
2.1 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 |
0.9 |
GO:0060737 |
prostate epithelial cord elongation(GO:0060523) prostate gland morphogenetic growth(GO:0060737) |
| 0.4 |
3.8 |
GO:0051150 |
regulation of smooth muscle cell differentiation(GO:0051150) |
| 0.4 |
5.9 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.4 |
11.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 |
2.0 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.4 |
0.4 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.4 |
2.0 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.4 |
0.8 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.4 |
1.2 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 |
0.8 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 |
1.9 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.4 |
3.1 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.4 |
1.5 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.4 |
6.0 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.4 |
1.9 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 |
1.5 |
GO:0000237 |
leptotene(GO:0000237) |
| 0.4 |
1.1 |
GO:1903519 |
negative regulation of interleukin-1 secretion(GO:0050711) negative regulation of interleukin-1 beta secretion(GO:0050713) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.4 |
1.1 |
GO:0060681 |
branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.4 |
2.9 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.4 |
1.4 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 |
0.7 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.3 |
1.7 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 |
1.0 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 |
1.4 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 |
2.0 |
GO:0045910 |
negative regulation of DNA recombination(GO:0045910) |
| 0.3 |
1.4 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 |
1.3 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.3 |
1.0 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.3 |
1.0 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 |
1.0 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.3 |
1.3 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 |
1.0 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 |
0.9 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.3 |
3.4 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.3 |
9.8 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 |
1.5 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.3 |
1.2 |
GO:0000733 |
DNA strand renaturation(GO:0000733) |
| 0.3 |
1.8 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 0.3 |
0.9 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.3 |
5.3 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.3 |
6.7 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.3 |
0.9 |
GO:1903722 |
endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) late endosomal microautophagy(GO:0061738) regulation of centriole elongation(GO:1903722) |
| 0.3 |
6.7 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.3 |
1.2 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 |
1.7 |
GO:0051461 |
corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 |
4.1 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.3 |
0.8 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.3 |
2.5 |
GO:0045719 |
negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 |
0.8 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.3 |
2.7 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 |
2.9 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.3 |
0.8 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.3 |
0.5 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) |
| 0.3 |
0.5 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.3 |
1.5 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 |
1.0 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.2 |
2.2 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.2 |
2.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.2 |
1.4 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.2 |
2.6 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.2 |
2.6 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 |
2.1 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 |
2.6 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.2 |
0.5 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.2 |
1.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.2 |
1.6 |
GO:0045915 |
positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.2 |
5.4 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 |
0.9 |
GO:1902224 |
cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.2 |
0.7 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
3.6 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.2 |
1.6 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.2 |
1.1 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.2 |
1.5 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 |
0.9 |
GO:0019230 |
proprioception(GO:0019230) |
| 0.2 |
5.9 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.2 |
8.5 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 |
0.6 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.2 |
0.6 |
GO:1901254 |
response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.2 |
1.0 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
3.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.2 |
0.4 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.2 |
1.6 |
GO:0048368 |
lateral mesoderm development(GO:0048368) |
| 0.2 |
0.8 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 |
1.2 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 |
0.6 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.2 |
1.5 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.2 |
0.9 |
GO:1903598 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.2 |
0.6 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
5.4 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.2 |
0.6 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 |
1.7 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.2 |
1.1 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 |
0.5 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.2 |
0.2 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 |
1.2 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 |
0.7 |
GO:0034239 |
regulation of macrophage fusion(GO:0034239) |
| 0.2 |
1.4 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 |
0.5 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.2 |
5.6 |
GO:0030071 |
regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.2 |
0.7 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.2 |
0.9 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 |
2.4 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 |
0.7 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.2 |
1.5 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.2 |
1.3 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.2 |
2.8 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 |
0.5 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.2 |
0.5 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 |
0.5 |
GO:0021987 |
cerebral cortex development(GO:0021987) |
| 0.2 |
0.5 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 |
1.7 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.1 |
1.5 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
1.3 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 |
0.4 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
1.6 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
1.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
1.0 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.1 |
2.8 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.1 |
1.4 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.1 |
0.4 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
1.0 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
1.6 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.1 |
1.9 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 |
1.1 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
2.5 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 |
0.7 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.5 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
0.9 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.7 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
1.4 |
GO:0034394 |
protein localization to cell surface(GO:0034394) |
| 0.1 |
0.3 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
0.9 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
0.4 |
GO:0045630 |
positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.1 |
1.4 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.1 |
1.8 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.9 |
GO:2000051 |
negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 |
5.9 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |
| 0.1 |
0.4 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 |
1.7 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.1 |
1.1 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 |
0.8 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 |
0.6 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
1.7 |
GO:0009191 |
ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 |
3.6 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
1.4 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.1 |
0.9 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.6 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.7 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.5 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
0.5 |
GO:2000269 |
regulation of fibroblast apoptotic process(GO:2000269) |
| 0.1 |
1.1 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.4 |
GO:0090024 |
negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 |
0.6 |
GO:0060916 |
mesenchymal cell proliferation involved in lung development(GO:0060916) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.1 |
1.4 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.7 |
GO:0019348 |
polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.1 |
0.5 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.1 |
10.5 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.1 |
3.7 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.1 |
0.5 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
0.9 |
GO:1902033 |
regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.1 |
1.1 |
GO:0003351 |
epithelial cilium movement(GO:0003351) |
| 0.1 |
1.0 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
0.6 |
GO:0033683 |
nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 |
0.6 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 |
2.4 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.6 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
1.7 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.1 |
1.6 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.1 |
2.1 |
GO:1902808 |
positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.1 |
1.3 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.1 |
1.0 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.5 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.6 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
0.6 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.5 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
4.3 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
5.0 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.1 |
3.0 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.1 |
0.4 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 |
1.8 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.1 |
1.2 |
GO:1902743 |
regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.1 |
0.4 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.8 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
1.5 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.1 |
0.5 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.1 |
0.6 |
GO:0015781 |
nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.1 |
0.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 |
0.3 |
GO:0009313 |
oligosaccharide catabolic process(GO:0009313) |
| 0.1 |
3.4 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.1 |
4.3 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.1 |
2.2 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 |
0.3 |
GO:0090325 |
negative regulation of multicellular organismal metabolic process(GO:0044252) regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 |
0.2 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 0.1 |
0.8 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 |
1.6 |
GO:0061028 |
establishment of endothelial barrier(GO:0061028) |
| 0.1 |
0.2 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 |
0.4 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 |
0.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.5 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
0.2 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.3 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
1.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.1 |
2.7 |
GO:0000070 |
mitotic sister chromatid segregation(GO:0000070) |
| 0.1 |
1.0 |
GO:0031498 |
nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.6 |
GO:0030224 |
monocyte differentiation(GO:0030224) |
| 0.1 |
0.4 |
GO:0032196 |
transposition(GO:0032196) |
| 0.1 |
3.5 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.1 |
1.2 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 |
0.6 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 |
0.4 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
0.5 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
0.7 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.1 |
0.4 |
GO:0006662 |
glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 |
1.2 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.1 |
0.1 |
GO:0030202 |
heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 |
3.6 |
GO:0051289 |
protein homotetramerization(GO:0051289) |
| 0.1 |
0.5 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.1 |
0.3 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
1.1 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 |
0.2 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 |
12.1 |
GO:0000398 |
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 |
1.8 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.1 |
0.7 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.1 |
0.2 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
2.7 |
GO:0032330 |
regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 |
0.6 |
GO:0036035 |
osteoclast development(GO:0036035) |
| 0.1 |
0.6 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
0.2 |
GO:0033089 |
positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 |
0.4 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.1 |
0.2 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.6 |
GO:0046686 |
response to cadmium ion(GO:0046686) |
| 0.1 |
0.5 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
3.2 |
GO:0009060 |
aerobic respiration(GO:0009060) |
| 0.1 |
0.3 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 |
1.1 |
GO:0002455 |
humoral immune response mediated by circulating immunoglobulin(GO:0002455) |
| 0.1 |
0.5 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.1 |
1.3 |
GO:0035315 |
hair cell differentiation(GO:0035315) |
| 0.1 |
0.2 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
0.1 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
0.1 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.8 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
1.0 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.0 |
0.1 |
GO:0061418 |
regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 |
0.7 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.2 |
GO:1901524 |
protein retention in Golgi apparatus(GO:0045053) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 |
0.3 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
1.7 |
GO:0031960 |
response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 |
0.2 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.5 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.2 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
1.0 |
GO:0019432 |
triglyceride biosynthetic process(GO:0019432) |
| 0.0 |
1.5 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.0 |
0.1 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.7 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 |
0.7 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.0 |
0.5 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
1.0 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.8 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
1.1 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.5 |
GO:0048712 |
negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 |
0.4 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.8 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.9 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.3 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.2 |
GO:0060271 |
cilium morphogenesis(GO:0060271) |
| 0.0 |
0.7 |
GO:0007249 |
I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 |
0.2 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.0 |
0.6 |
GO:0001779 |
natural killer cell differentiation(GO:0001779) |
| 0.0 |
0.6 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
1.5 |
GO:0042274 |
ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 |
0.9 |
GO:0035904 |
aorta development(GO:0035904) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.3 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
1.1 |
GO:0002011 |
morphogenesis of an epithelial sheet(GO:0002011) |
| 0.0 |
1.3 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
1.2 |
GO:0001959 |
regulation of cytokine-mediated signaling pathway(GO:0001959) |
| 0.0 |
1.7 |
GO:0017158 |
regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 |
0.2 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.5 |
GO:0048661 |
positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 |
0.6 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.3 |
GO:0001947 |
heart looping(GO:0001947) embryonic heart tube morphogenesis(GO:0003143) determination of heart left/right asymmetry(GO:0061371) |
| 0.0 |
0.5 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.2 |
GO:0000466 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) cleavage involved in rRNA processing(GO:0000469) |
| 0.0 |
0.9 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.2 |
GO:0007398 |
ectoderm development(GO:0007398) |
| 0.0 |
0.1 |
GO:1904714 |
chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
1.2 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0060406 |
positive regulation of penile erection(GO:0060406) |
| 0.0 |
0.2 |
GO:1903035 |
negative regulation of response to wounding(GO:1903035) |
| 0.0 |
0.7 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.5 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.2 |
GO:0045589 |
regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.5 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.1 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.2 |
GO:0060213 |
positive regulation of mRNA 3'-end processing(GO:0031442) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
1.2 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.2 |
GO:0032402 |
melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.2 |
GO:0022613 |
ribonucleoprotein complex biogenesis(GO:0022613) |
| 0.0 |
0.1 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
0.2 |
GO:0042832 |
defense response to protozoan(GO:0042832) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.1 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.0 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 |
0.2 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
0.5 |
GO:0006635 |
fatty acid beta-oxidation(GO:0006635) |