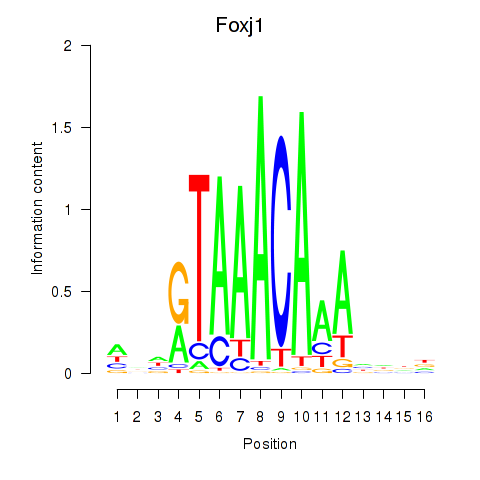
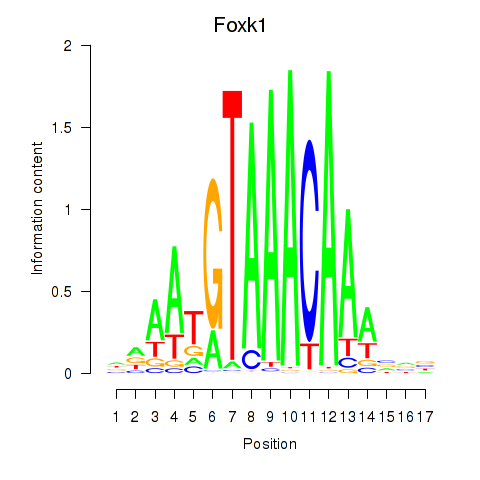
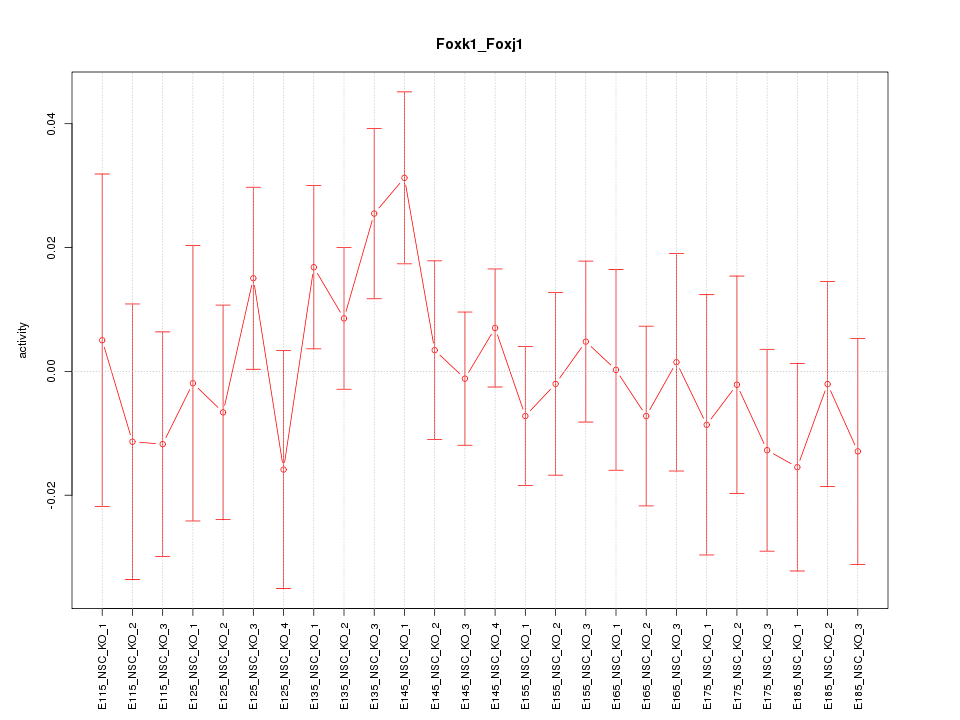
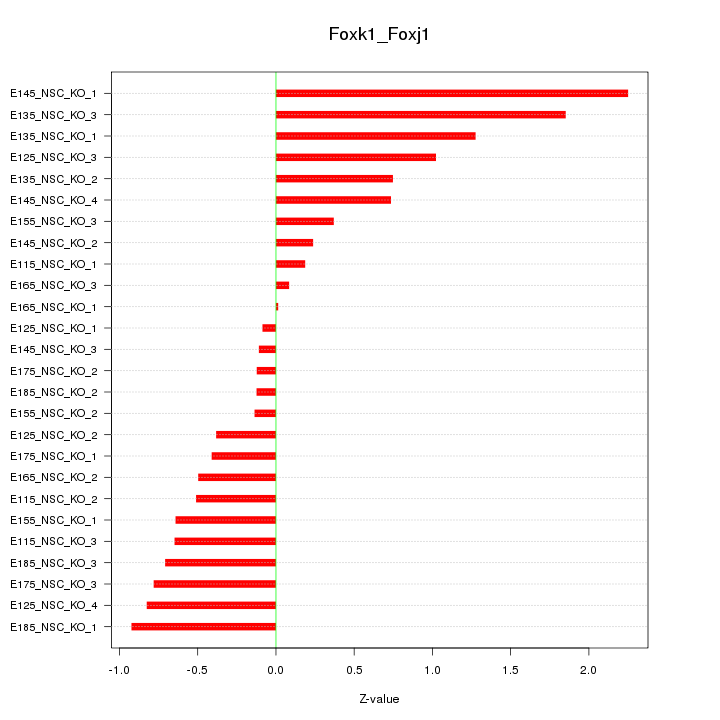
| 0.3 |
1.0 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 |
1.2 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
0.8 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 |
0.8 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 |
0.9 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 |
0.5 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 |
0.5 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 |
0.5 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 |
0.6 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 |
0.6 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.4 |
GO:0072139 |
glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 |
0.4 |
GO:1903061 |
regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.1 |
0.5 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
1.6 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.7 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
1.0 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
0.4 |
GO:0051311 |
meiotic metaphase plate congression(GO:0051311) |
| 0.1 |
0.5 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
1.4 |
GO:0071459 |
protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 |
0.3 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.1 |
0.2 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.4 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 |
0.4 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.1 |
0.2 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.1 |
0.4 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.2 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.5 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 |
0.3 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
0.2 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
0.7 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 |
0.3 |
GO:0035234 |
ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 |
0.2 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.1 |
GO:2000670 |
positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 |
1.1 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
0.2 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.3 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.0 |
0.2 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.5 |
GO:0030836 |
positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 |
0.2 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
2.4 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.2 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 |
0.2 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.0 |
0.2 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.0 |
0.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.7 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 |
0.2 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.2 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.0 |
0.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 |
0.2 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.8 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
0.2 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.0 |
0.1 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.7 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.1 |
GO:2000834 |
luteinizing hormone secretion(GO:0032275) androgen secretion(GO:0035935) testosterone secretion(GO:0035936) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 |
0.1 |
GO:0042633 |
molting cycle(GO:0042303) hair cycle(GO:0042633) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0006545 |
glycine biosynthetic process(GO:0006545) |
| 0.0 |
0.1 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.8 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.1 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.1 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 |
0.4 |
GO:0042640 |
anagen(GO:0042640) |
| 0.0 |
0.2 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.1 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.1 |
GO:0010616 |
negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of protein glycosylation(GO:0060051) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.1 |
GO:0010693 |
negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.2 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 |
0.1 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.1 |
GO:1902071 |
regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 |
0.1 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.0 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
0.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
0.1 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 |
0.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.1 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.3 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.1 |
GO:0015817 |
histidine transport(GO:0015817) |
| 0.0 |
0.1 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.0 |
0.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.5 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.0 |
0.1 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0048318 |
axial mesoderm development(GO:0048318) |
| 0.0 |
0.0 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.0 |
0.1 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.1 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.0 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.4 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.0 |
0.2 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.0 |
0.5 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.4 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.2 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.0 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 |
0.3 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |