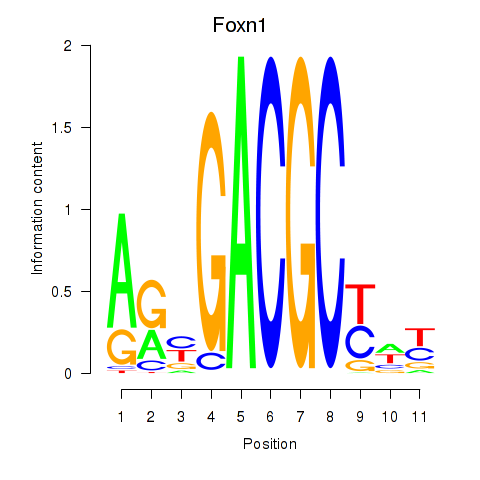
| 0.2 |
0.6 |
GO:0086017 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) Purkinje myocyte action potential(GO:0086017) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 |
1.2 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.1 |
0.4 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.3 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.2 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.1 |
0.2 |
GO:0071034 |
CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 |
0.3 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.4 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 |
0.2 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 |
0.2 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.0 |
0.2 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.1 |
GO:0001828 |
inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 |
0.1 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.0 |
0.2 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.1 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
0.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.2 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 |
0.2 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.4 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
0.0 |
GO:1903061 |
glutamate secretion, neurotransmission(GO:0061535) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.0 |
0.1 |
GO:0035845 |
photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 |
0.2 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.1 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.0 |
0.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.1 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.2 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.0 |
0.2 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.0 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.0 |
0.0 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.0 |
0.3 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.0 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |