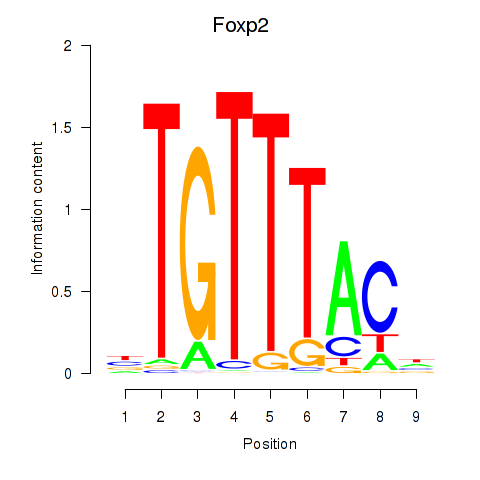
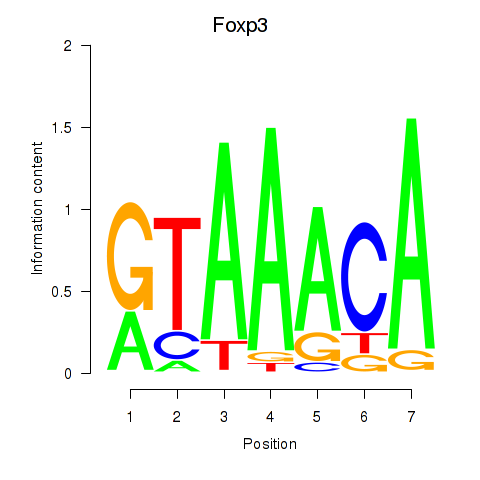
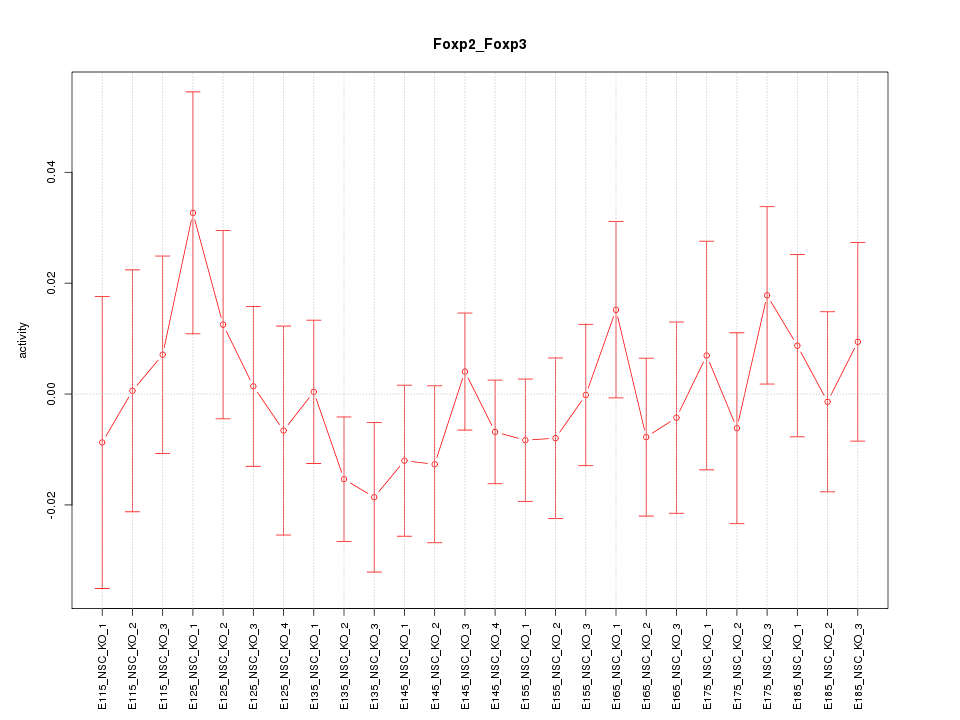
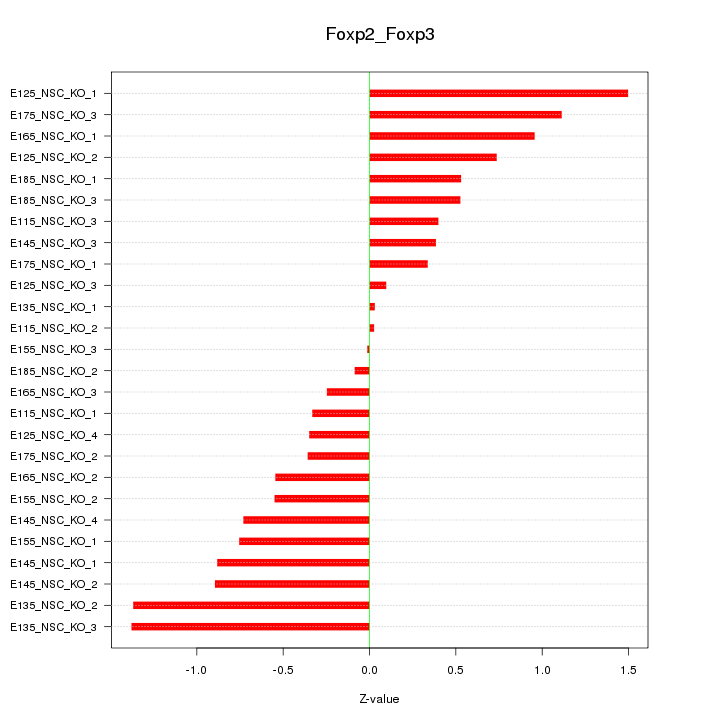
| 1.0 |
2.9 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.6 |
1.7 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.5 |
1.5 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.4 |
1.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.3 |
2.4 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.3 |
0.9 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.3 |
0.9 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.3 |
2.3 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.3 |
0.8 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 |
0.6 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
3.7 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.2 |
0.6 |
GO:0072048 |
pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.2 |
0.6 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 |
1.7 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 |
0.6 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.2 |
1.0 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 |
0.8 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.2 |
0.9 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.2 |
0.7 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
0.5 |
GO:1902524 |
negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.2 |
2.1 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.2 |
0.5 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.2 |
3.2 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 |
0.5 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 |
0.8 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.2 |
0.7 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.2 |
1.0 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.2 |
0.5 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 |
1.2 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.2 |
0.8 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
0.1 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.4 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.1 |
0.4 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.1 |
0.7 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
1.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.5 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
0.9 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 |
2.2 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
1.6 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.8 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.1 |
0.4 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.1 |
0.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.9 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.3 |
GO:2000598 |
regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.1 |
0.4 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 |
0.4 |
GO:0070236 |
negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 |
0.3 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.3 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 |
0.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.3 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 |
0.2 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.6 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
0.4 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.2 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 |
0.1 |
GO:2000078 |
positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.1 |
0.2 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.1 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.1 |
0.8 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.3 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
0.9 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.9 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.5 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
0.4 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.3 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 |
0.4 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.3 |
GO:2000973 |
regulation of pro-B cell differentiation(GO:2000973) |
| 0.1 |
1.0 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
0.2 |
GO:0051088 |
PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 |
0.4 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
1.3 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 |
0.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.3 |
GO:0008655 |
pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.9 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.1 |
0.4 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.4 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
0.4 |
GO:0060872 |
semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 |
1.2 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
1.9 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.3 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.8 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.5 |
GO:1901621 |
regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.2 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.0 |
0.4 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.0 |
0.1 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.4 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.1 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.2 |
GO:0071105 |
negative regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003340) response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 |
0.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.1 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.6 |
GO:0071542 |
dopaminergic neuron differentiation(GO:0071542) |
| 0.0 |
0.3 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.5 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.2 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.0 |
0.3 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.0 |
1.2 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.2 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.1 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.0 |
0.1 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.0 |
0.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.0 |
0.1 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.2 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.0 |
0.1 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.2 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.5 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.5 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.0 |
0.1 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.4 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.1 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.4 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
1.1 |
GO:0046326 |
positive regulation of glucose import(GO:0046326) |
| 0.0 |
0.2 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.3 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.2 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.9 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
0.1 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.4 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:0002347 |
response to tumor cell(GO:0002347) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.2 |
GO:0015868 |
purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 |
1.5 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.2 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.1 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.0 |
0.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.1 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.3 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.1 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.0 |
0.1 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.4 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
0.1 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.1 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.1 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.2 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.1 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.2 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.7 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 |
0.2 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.1 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.1 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.1 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |