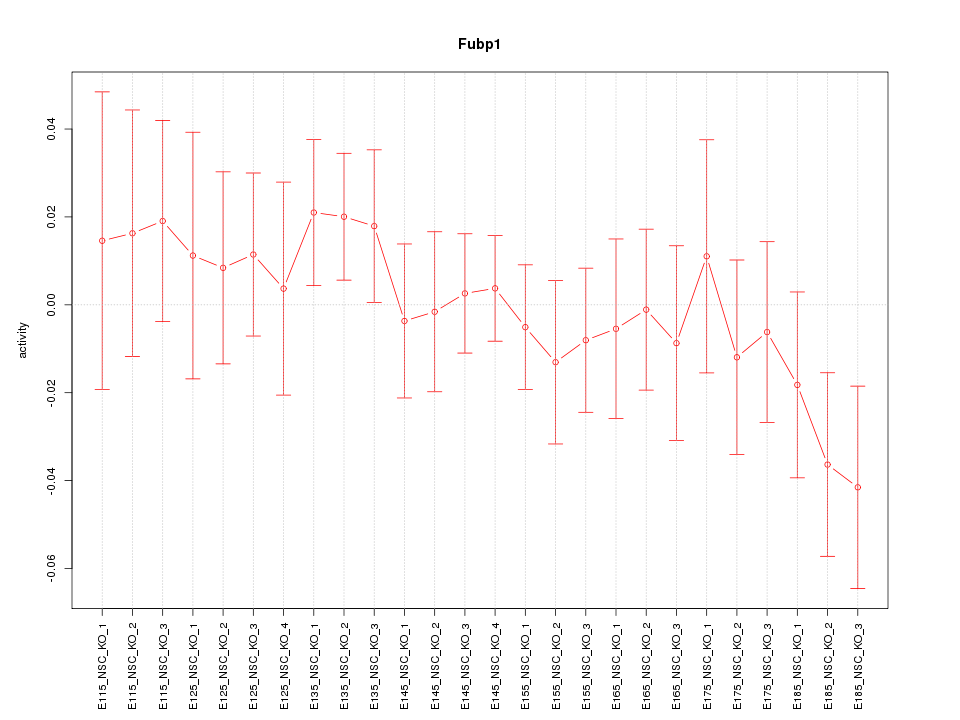
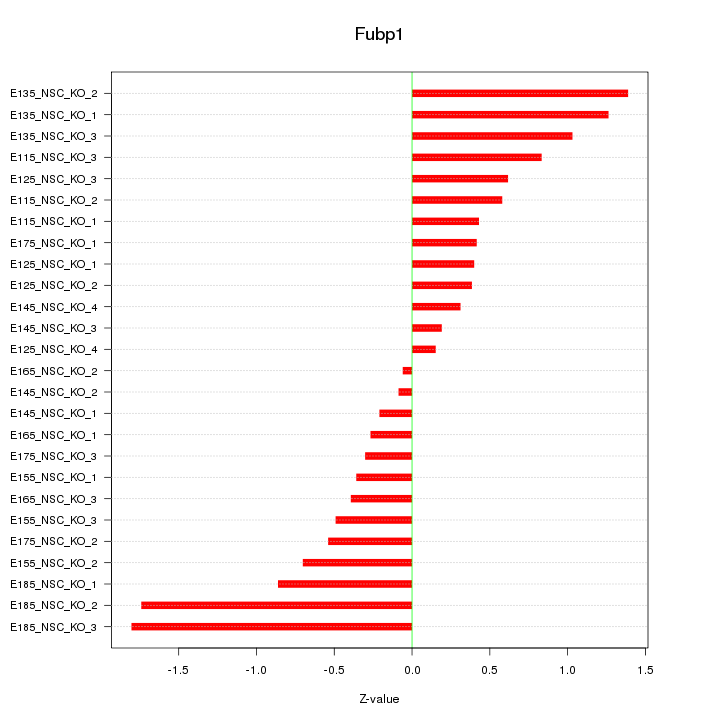
| 0.8 |
3.1 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.7 |
2.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.7 |
8.6 |
GO:0097067 |
negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.5 |
2.0 |
GO:0098763 |
mitotic cell cycle phase(GO:0098763) |
| 0.5 |
1.0 |
GO:0046985 |
positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.5 |
1.0 |
GO:1905065 |
positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.4 |
1.2 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.3 |
0.6 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 |
0.8 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.2 |
1.2 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.2 |
0.6 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.2 |
0.6 |
GO:0060129 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.2 |
2.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.2 |
0.5 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.4 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.1 |
0.4 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 |
0.4 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
0.4 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 |
0.9 |
GO:1903301 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.5 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.1 |
0.3 |
GO:0046881 |
sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 |
0.6 |
GO:2000002 |
negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 |
1.0 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.5 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 |
0.3 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.4 |
GO:0048133 |
NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.4 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.1 |
0.5 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
0.7 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 |
0.3 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.6 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
2.3 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.0 |
0.1 |
GO:1902336 |
neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) regulation of retinal ganglion cell axon guidance(GO:0090259) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.0 |
0.2 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.1 |
GO:0050883 |
musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 |
0.5 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.2 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.7 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
2.4 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 |
0.4 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.0 |
0.6 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.7 |
GO:0042493 |
response to drug(GO:0042493) |
| 0.0 |
0.3 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.2 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.1 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 |
1.7 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 0.0 |
0.5 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.6 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.3 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.3 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.2 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.1 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.3 |
GO:0070266 |
necroptotic process(GO:0070266) |