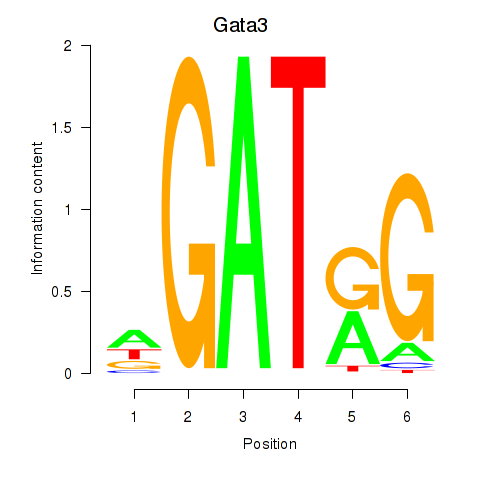
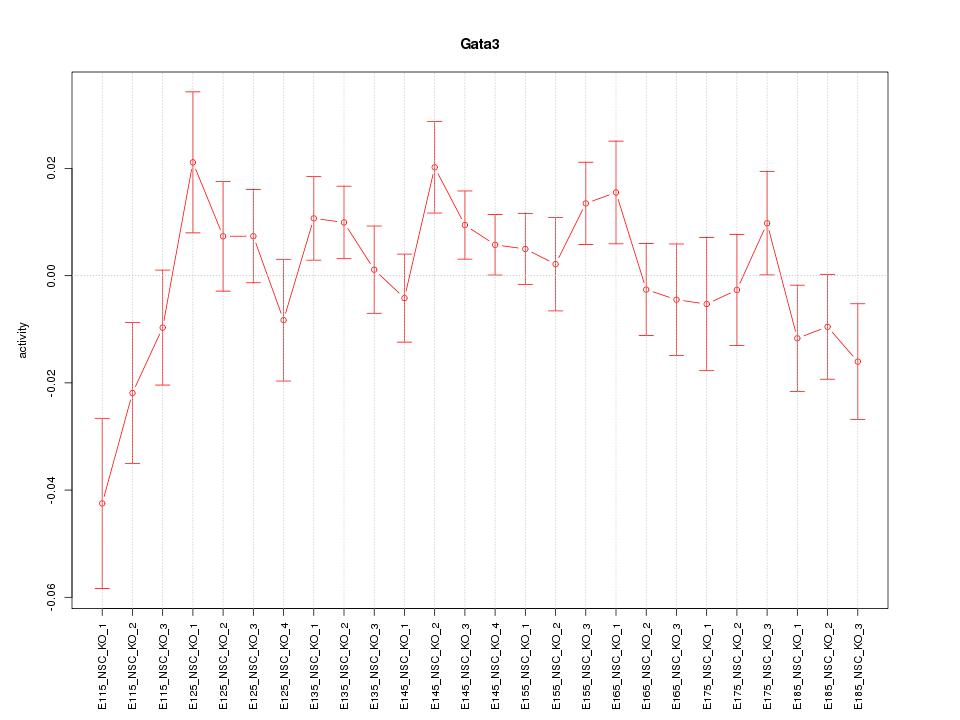
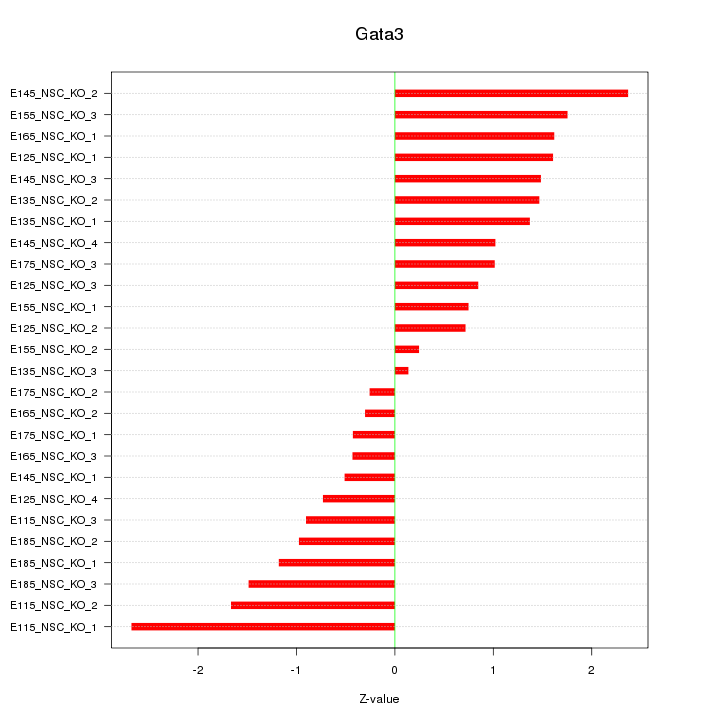
| 0.8 |
3.3 |
GO:0007412 |
axon target recognition(GO:0007412) |
| 0.7 |
2.0 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.6 |
2.9 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.5 |
3.3 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.5 |
1.6 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 |
2.1 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 |
2.7 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.5 |
1.9 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.4 |
1.6 |
GO:0002175 |
protein localization to paranode region of axon(GO:0002175) |
| 0.4 |
1.6 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.4 |
2.4 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.4 |
1.2 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 |
1.5 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 |
2.6 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.4 |
1.8 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.4 |
2.8 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 |
1.4 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 |
1.3 |
GO:0061030 |
epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.3 |
1.9 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 |
4.8 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 |
1.8 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 |
2.8 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 |
1.3 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.3 |
0.8 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.3 |
0.8 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 |
0.5 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.3 |
0.8 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.2 |
1.5 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.2 |
0.7 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.2 |
0.7 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.2 |
0.7 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 |
1.2 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.2 |
2.3 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 |
0.7 |
GO:0032829 |
positive regulation of T cell anergy(GO:0002669) negative regulation of chronic inflammatory response(GO:0002677) positive regulation of lymphocyte anergy(GO:0002913) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) negative regulation of interleukin-2 biosynthetic process(GO:0045085) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 |
1.5 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 |
0.9 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
5.0 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.2 |
0.9 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 |
1.5 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.2 |
0.9 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.2 |
1.1 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.2 |
0.2 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) |
| 0.2 |
1.0 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.2 |
0.6 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.2 |
2.7 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.2 |
0.8 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.2 |
1.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.2 |
0.5 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 |
0.5 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.2 |
0.5 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.2 |
0.5 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 |
1.2 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.2 |
1.1 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 |
0.8 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.2 |
0.8 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 |
0.7 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 |
0.6 |
GO:0046958 |
nonassociative learning(GO:0046958) |
| 0.1 |
0.8 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 |
1.3 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.4 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.1 |
0.4 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.1 |
1.0 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.1 |
0.5 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 |
1.1 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.1 |
0.5 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 |
0.5 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
0.5 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.6 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
2.2 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.1 |
0.4 |
GO:0016115 |
terpenoid catabolic process(GO:0016115) |
| 0.1 |
0.4 |
GO:0009223 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 |
1.9 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.4 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.8 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 |
1.1 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.1 |
0.6 |
GO:0019530 |
taurine metabolic process(GO:0019530) |
| 0.1 |
0.9 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
0.3 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
2.5 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 |
0.7 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.6 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 |
0.3 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
4.4 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.1 |
0.3 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.4 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 |
0.4 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 |
1.5 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.4 |
GO:0055099 |
response to high density lipoprotein particle(GO:0055099) |
| 0.1 |
0.3 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.9 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
0.9 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.6 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
1.1 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.1 |
0.6 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.1 |
0.6 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.1 |
0.4 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 |
0.5 |
GO:0061001 |
regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 |
0.5 |
GO:0032329 |
serine transport(GO:0032329) |
| 0.1 |
1.0 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
0.3 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 |
0.6 |
GO:0038171 |
cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.4 |
GO:1903598 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) positive regulation of gap junction assembly(GO:1903598) |
| 0.1 |
0.3 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
0.3 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
1.6 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.1 |
0.2 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.1 |
0.2 |
GO:0031038 |
meiotic chromosome movement towards spindle pole(GO:0016344) myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.5 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.1 |
1.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.5 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.1 |
0.6 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.2 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.1 |
0.7 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.1 |
0.8 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
0.7 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 |
0.3 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 |
1.7 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.2 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.1 |
0.4 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
0.2 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
0.4 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.1 |
0.3 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
0.2 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.4 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 |
0.4 |
GO:1903056 |
mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.3 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
0.2 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.4 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 |
0.3 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
0.2 |
GO:0070488 |
regulation of integrin biosynthetic process(GO:0045113) neutrophil aggregation(GO:0070488) |
| 0.1 |
1.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.1 |
0.8 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.4 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
0.7 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.3 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
0.6 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.7 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
1.8 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.1 |
6.0 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.1 |
0.9 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.4 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 |
0.2 |
GO:0002086 |
diaphragm contraction(GO:0002086) |
| 0.1 |
0.4 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.1 |
1.4 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.1 |
0.5 |
GO:0006977 |
DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 |
0.7 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.2 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.2 |
GO:0046880 |
gonadotropin secretion(GO:0032274) regulation of gonadotropin secretion(GO:0032276) regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.1 |
0.7 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
0.6 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.5 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.4 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 |
0.4 |
GO:2000650 |
negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.1 |
GO:0071384 |
cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 |
0.2 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.0 |
0.2 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
1.0 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 |
2.4 |
GO:0042073 |
intraciliary transport(GO:0042073) |
| 0.0 |
0.2 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.4 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 |
0.3 |
GO:0048537 |
mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
1.5 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.7 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.0 |
0.7 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.2 |
GO:1901096 |
regulation of autophagosome maturation(GO:1901096) |
| 0.0 |
1.9 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.2 |
GO:2000152 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.7 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.5 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 |
0.1 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.2 |
GO:2001286 |
regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 |
0.1 |
GO:1900426 |
positive regulation of defense response to bacterium(GO:1900426) |
| 0.0 |
1.3 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.2 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.0 |
0.4 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.2 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.2 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.0 |
0.3 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 |
0.7 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 |
2.9 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.1 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 |
0.3 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
0.6 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
2.2 |
GO:0007612 |
learning(GO:0007612) |
| 0.0 |
0.9 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 |
1.2 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.9 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.8 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.4 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.0 |
0.5 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.8 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
1.1 |
GO:0050775 |
positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 |
0.3 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.3 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.6 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.2 |
GO:0000103 |
sulfate assimilation(GO:0000103) |
| 0.0 |
0.6 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.3 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
1.0 |
GO:0000460 |
maturation of 5.8S rRNA(GO:0000460) |
| 0.0 |
0.6 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
0.1 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 |
0.6 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.2 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.0 |
0.3 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.2 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.0 |
0.1 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 |
0.5 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 |
0.3 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 |
0.0 |
GO:0001844 |
protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 |
0.5 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.4 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.8 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.3 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 |
0.1 |
GO:0038083 |
peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 |
1.0 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.1 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 |
0.2 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.2 |
GO:0070242 |
thymocyte apoptotic process(GO:0070242) |
| 0.0 |
0.4 |
GO:0007263 |
nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.3 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.3 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.0 |
0.2 |
GO:0048169 |
regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 |
0.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.0 |
0.2 |
GO:0032367 |
intracellular cholesterol transport(GO:0032367) |
| 0.0 |
0.3 |
GO:0000303 |
response to superoxide(GO:0000303) |
| 0.0 |
0.1 |
GO:0014809 |
regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 |
0.3 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.1 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.1 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
1.1 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
1.2 |
GO:0007613 |
memory(GO:0007613) |
| 0.0 |
1.2 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.1 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.5 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.2 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
1.6 |
GO:0051101 |
regulation of DNA binding(GO:0051101) |
| 0.0 |
0.6 |
GO:0045761 |
regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 |
0.5 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.3 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
1.0 |
GO:0031110 |
regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 |
0.2 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.1 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 |
0.1 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.0 |
0.5 |
GO:0048662 |
negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 |
0.3 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.4 |
GO:0048168 |
regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 |
0.2 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 |
0.4 |
GO:0048247 |
lymphocyte chemotaxis(GO:0048247) |
| 0.0 |
0.2 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.0 |
0.4 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.8 |
GO:0016079 |
synaptic vesicle exocytosis(GO:0016079) |
| 0.0 |
0.1 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.8 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.0 |
0.1 |
GO:0010226 |
response to lithium ion(GO:0010226) |
| 0.0 |
0.1 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.2 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.8 |
GO:0050890 |
cognition(GO:0050890) |
| 0.0 |
0.4 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.2 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.4 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.0 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.0 |
0.6 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.1 |
GO:0032691 |
negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.1 |
GO:0046322 |
negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 |
0.3 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.3 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.0 |
GO:0070625 |
zymogen granule exocytosis(GO:0070625) |
| 0.0 |
0.1 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.2 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.0 |
1.2 |
GO:0008643 |
carbohydrate transport(GO:0008643) |
| 0.0 |
0.2 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.2 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.3 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
1.0 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.2 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.1 |
GO:0002093 |
auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
1.1 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.1 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |