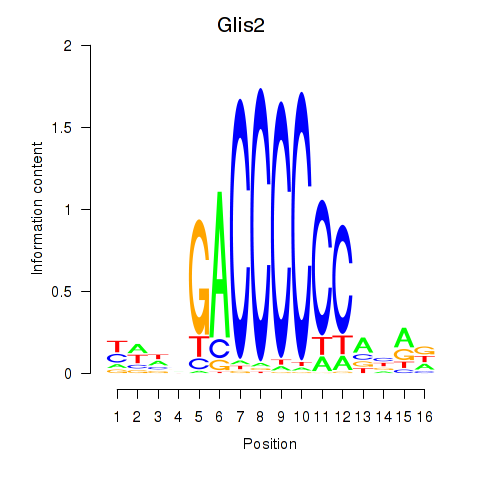
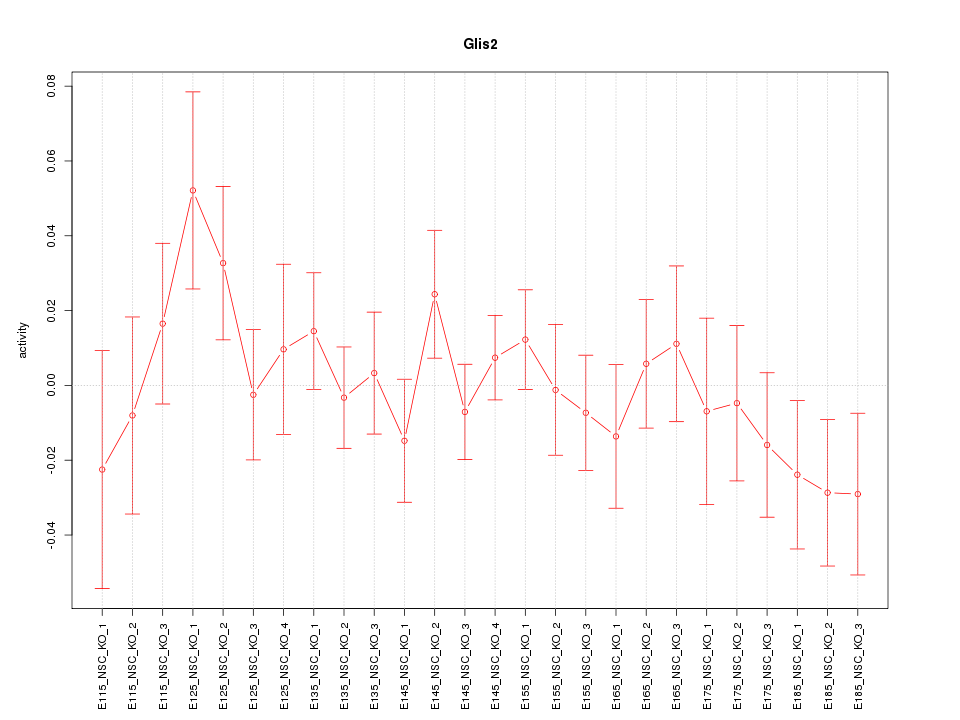
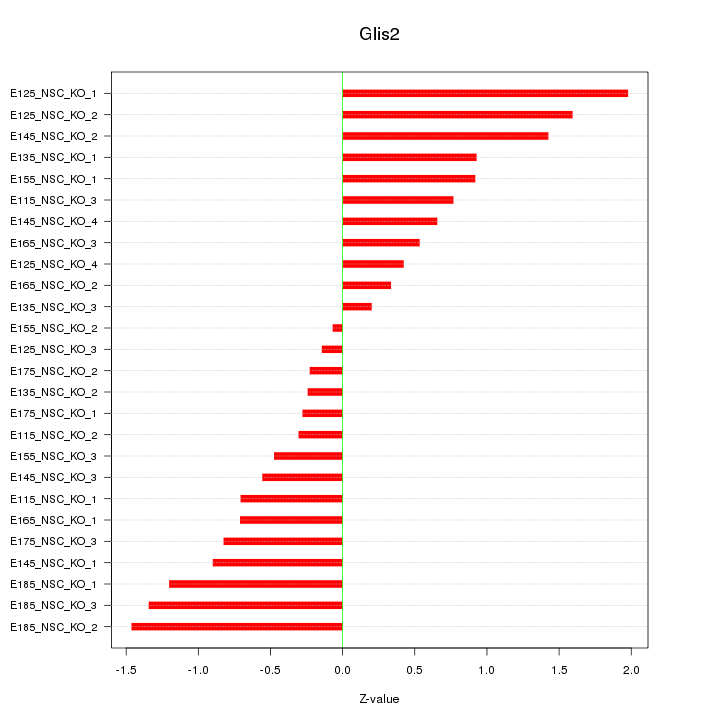
| 0.9 |
2.8 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.7 |
6.6 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.6 |
2.4 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.5 |
3.7 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.5 |
3.7 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.4 |
2.1 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.4 |
5.0 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.3 |
1.8 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.2 |
0.7 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.2 |
0.9 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) retinal rod cell differentiation(GO:0060221) |
| 0.2 |
0.6 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 |
1.0 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.2 |
0.5 |
GO:0010899 |
regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.2 |
0.5 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.2 |
0.8 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.6 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.6 |
GO:2000054 |
negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 |
0.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
0.6 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
0.9 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.5 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.1 |
0.3 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.1 |
1.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.5 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.1 |
0.4 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 |
0.5 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.1 |
0.7 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.9 |
GO:0045040 |
protein import into mitochondrial outer membrane(GO:0045040) negative regulation of protein folding(GO:1903333) |
| 0.1 |
0.3 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
0.8 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.3 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.8 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.1 |
3.0 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.4 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
0.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
8.3 |
GO:0007498 |
mesoderm development(GO:0007498) |
| 0.1 |
0.5 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.3 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.1 |
0.2 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.1 |
0.4 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.8 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.3 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.6 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
0.3 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.4 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.1 |
1.4 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.5 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.0 |
0.2 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.5 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.4 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.3 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 |
0.3 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 |
0.4 |
GO:0035729 |
response to hepatocyte growth factor(GO:0035728) cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 |
0.5 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.0 |
0.3 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.4 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.2 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.3 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.6 |
GO:0042297 |
vocal learning(GO:0042297) imitative learning(GO:0098596) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.0 |
0.6 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
0.8 |
GO:0048025 |
negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 |
0.2 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.2 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.6 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0015697 |
quaternary ammonium group transport(GO:0015697) |
| 0.0 |
0.2 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.1 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.0 |
0.4 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.7 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.7 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.6 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.2 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.3 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.1 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.4 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 |
0.5 |
GO:0043303 |
mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 |
0.3 |
GO:0042921 |
glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.1 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.5 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.3 |
GO:0006221 |
pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 |
0.2 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.1 |
GO:0021592 |
fourth ventricle development(GO:0021592) initiation of neural tube closure(GO:0021993) |