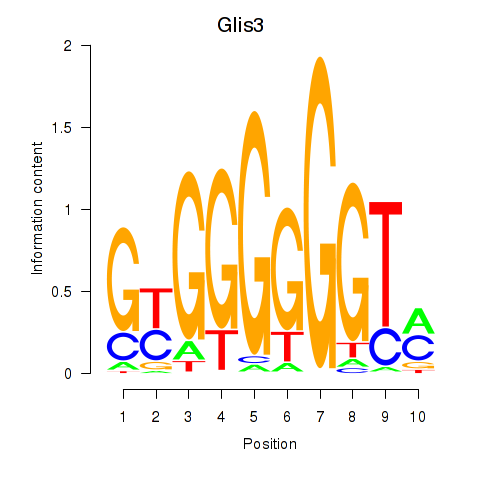
| 0.5 |
1.6 |
GO:0010725 |
regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.4 |
1.1 |
GO:0070172 |
oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.3 |
1.3 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.3 |
2.8 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 |
1.9 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
0.6 |
GO:0003032 |
detection of oxygen(GO:0003032) |
| 0.1 |
0.6 |
GO:0097374 |
sensory neuron axon guidance(GO:0097374) |
| 0.1 |
0.7 |
GO:0048133 |
NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
1.2 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
2.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.7 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.1 |
0.8 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.4 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.1 |
0.2 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.1 |
0.3 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.2 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.2 |
GO:0003100 |
angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) regulation of systemic arterial blood pressure by endothelin(GO:0003100) beta selection(GO:0043366) |
| 0.0 |
0.4 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
0.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.3 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.3 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.0 |
0.1 |
GO:0048146 |
regulation of fibroblast proliferation(GO:0048145) positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 |
0.6 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.2 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 |
0.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.8 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.2 |
GO:0001771 |
immunological synapse formation(GO:0001771) |
| 0.0 |
0.1 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 |
0.2 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.1 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 |
0.2 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.1 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.0 |
0.1 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.0 |
0.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.3 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.2 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.4 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.0 |
0.2 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.3 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.1 |
GO:0032811 |
negative regulation of epinephrine secretion(GO:0032811) |
| 0.0 |
0.5 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.0 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.0 |
0.2 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.2 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.0 |
0.3 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.4 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |