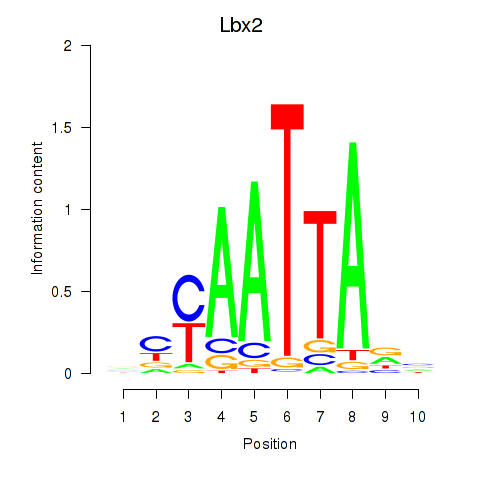
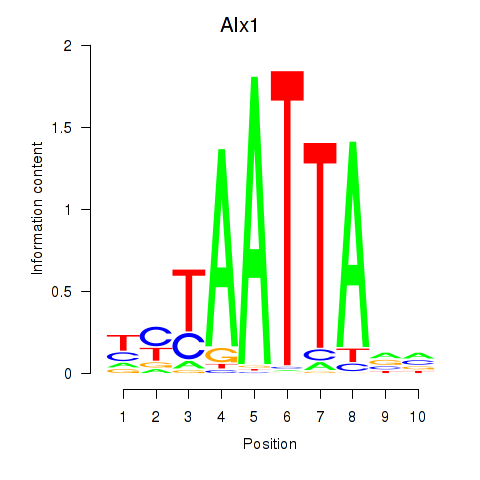
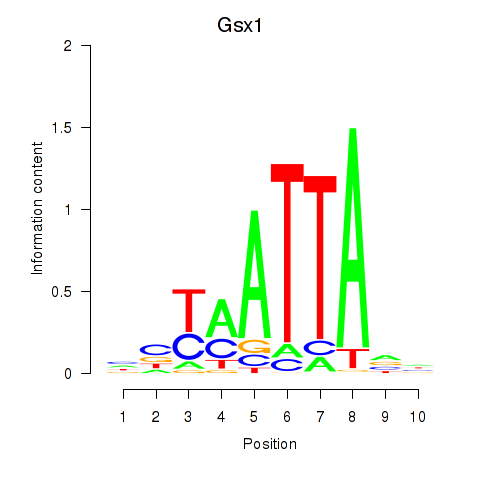
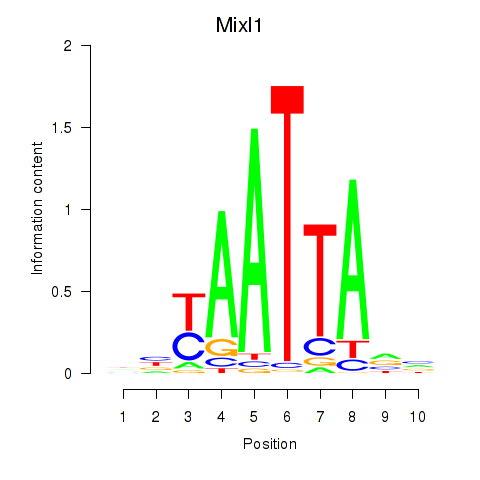
| 0.9 |
2.8 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 |
1.9 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 |
1.2 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.3 |
0.9 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 |
2.9 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 |
0.7 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.2 |
0.7 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.2 |
0.9 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.2 |
1.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
0.8 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 |
1.5 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 |
0.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
1.0 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.1 |
1.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
1.1 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.8 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
0.5 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
0.4 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 |
0.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.7 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
1.8 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.7 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.2 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 |
0.2 |
GO:0035790 |
platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 |
0.5 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 |
0.2 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 |
0.4 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.1 |
1.0 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.7 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 |
0.1 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.1 |
0.3 |
GO:0070814 |
hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 |
0.2 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.2 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.3 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.1 |
GO:0045852 |
pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 |
0.3 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.5 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.2 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 |
1.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.4 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.7 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.1 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.0 |
0.3 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.6 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.7 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.3 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
0.2 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.1 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.2 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 |
0.2 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.0 |
0.1 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 |
0.1 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.0 |
0.1 |
GO:0090298 |
negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.0 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.3 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
1.0 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.1 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.1 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 |
0.1 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.1 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
1.2 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.5 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.0 |
0.4 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
1.1 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
4.1 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.2 |
GO:0032196 |
transposition(GO:0032196) |
| 0.0 |
0.8 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.9 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.1 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 |
1.8 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.5 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.3 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.6 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.5 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
0.1 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 |
0.3 |
GO:1901673 |
regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 |
0.1 |
GO:0034397 |
telomere localization(GO:0034397) |
| 0.0 |
0.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.3 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.1 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.2 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.3 |
GO:0042994 |
cytoplasmic sequestering of transcription factor(GO:0042994) |
| 0.0 |
0.1 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 |
0.1 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.1 |
GO:0060768 |
epithelial cell proliferation involved in prostate gland development(GO:0060767) regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.1 |
GO:0044828 |
negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 |
0.2 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.1 |
GO:1902894 |
negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 |
0.1 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.2 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.3 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.5 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.2 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.1 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.4 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 |
0.2 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |