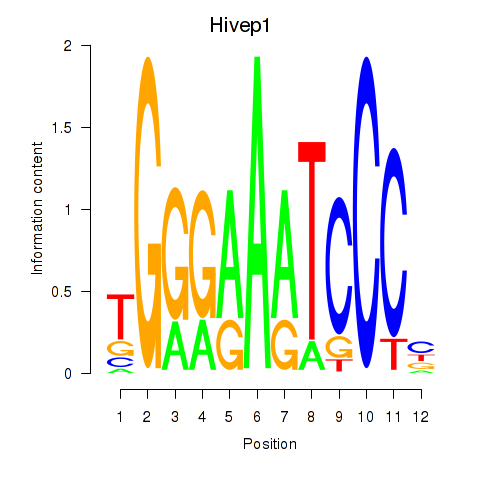
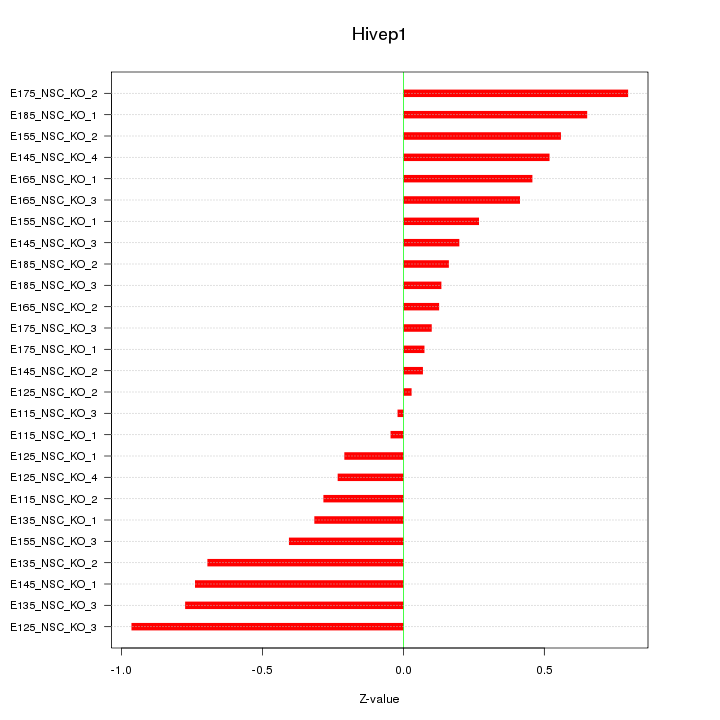
| 0.3 |
1.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.3 |
2.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.2 |
1.1 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.2 |
0.6 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 |
0.6 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.2 |
1.1 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
0.5 |
GO:1901228 |
regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 |
0.3 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.2 |
1.8 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.1 |
0.4 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.7 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 |
0.4 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.3 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 |
0.4 |
GO:0061428 |
positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
1.0 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.1 |
0.6 |
GO:0032713 |
negative regulation of interleukin-4 production(GO:0032713) |
| 0.1 |
0.6 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.1 |
0.3 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 |
0.6 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
1.3 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.5 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
0.3 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.1 |
0.2 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.3 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 |
0.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.1 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 |
0.6 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 |
0.5 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.0 |
1.1 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.1 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.0 |
0.6 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.0 |
0.1 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.6 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
1.0 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.2 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.3 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) positive regulation of vesicle fusion(GO:0031340) positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
1.2 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.3 |
GO:0032688 |
negative regulation of interferon-beta production(GO:0032688) |
| 0.0 |
1.8 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.4 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.1 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.0 |
0.1 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.3 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.1 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.3 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |