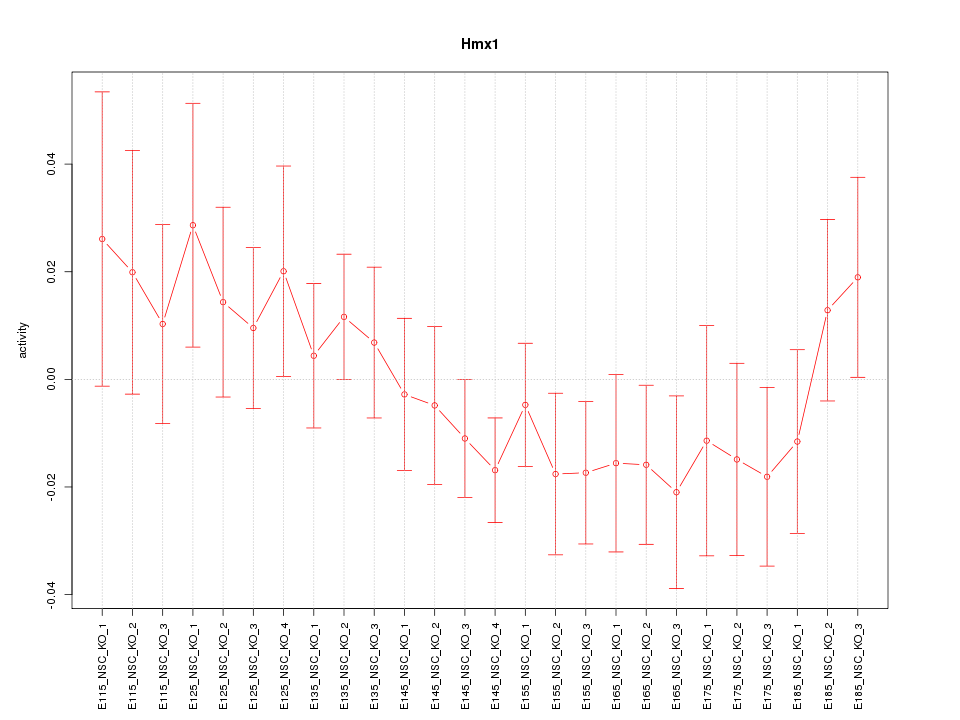
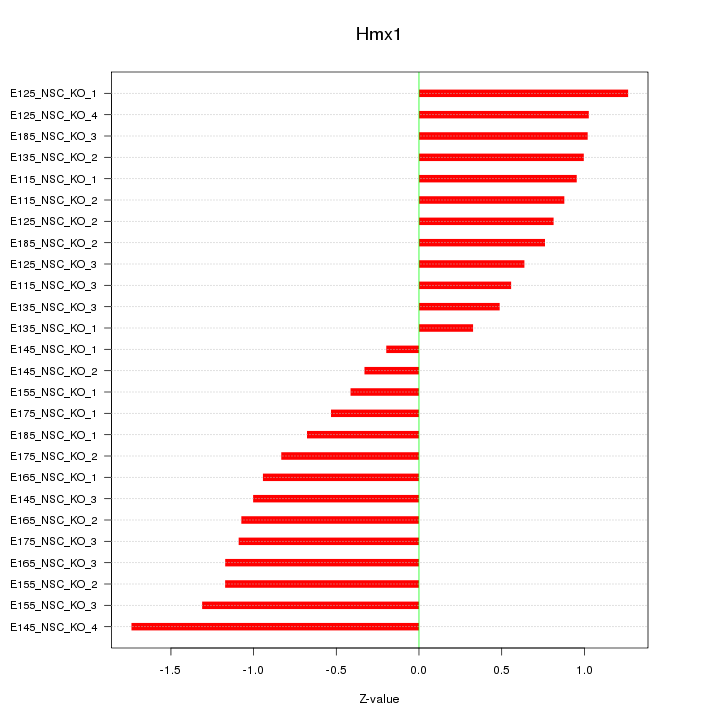
Motif ID: Hmx1
Z-value: 0.923

Transcription factors associated with Hmx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hmx1 | ENSMUSG00000067438.3 | Hmx1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 1.3 | 3.8 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.0 | 4.1 | GO:0032439 | endosome localization(GO:0032439) |
| 1.0 | 2.9 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.8 | 3.3 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.8 | 5.3 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.5 | 6.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 1.9 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.5 | 2.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.5 | 1.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 | 1.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.4 | 1.3 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.4 | 3.6 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.3 | 1.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.3 | 1.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 | 1.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.3 | 0.9 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.3 | 1.4 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 2.5 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.3 | 0.8 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 | 0.8 | GO:0014735 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) regulation of muscle atrophy(GO:0014735) |
| 0.3 | 0.8 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.3 | 1.0 | GO:0015744 | succinate transport(GO:0015744) |
| 0.2 | 3.9 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 1.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 3.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 1.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.2 | 2.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 1.9 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.2 | 1.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.8 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 0.2 | 1.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 2.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 4.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 3.7 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 0.5 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.7 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 2.5 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 1.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.1 | 1.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.9 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.8 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.6 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 | 0.5 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.1 | 0.5 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.2 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.5 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 2.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.2 | GO:0033523 | endodermal cell fate commitment(GO:0001711) histone H2B ubiquitination(GO:0033523) |
| 0.1 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 0.6 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.0 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.1 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 | 1.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 1.0 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.3 | GO:1902669 | positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.1 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.4 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.3 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.0 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.5 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.8 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 5.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 1.1 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:0032382 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) regulation of cholesterol import(GO:0060620) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) regulation of sterol import(GO:2000909) positive regulation of sterol import(GO:2000911) |
| 0.0 | 0.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.5 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.6 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.8 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 1.4 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.3 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 1.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 3.5 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.2 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 2.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 1.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 1.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.4 | 2.5 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.3 | 1.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 2.3 | GO:0005775 | vacuolar lumen(GO:0005775) |
| 0.3 | 1.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 2.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 2.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 3.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 7.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 0.8 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 1.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 6.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 1.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 6.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 7.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 3.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 0.8 | 3.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 1.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.3 | 0.9 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.3 | 2.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 1.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.3 | 1.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 2.5 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.2 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 5.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 3.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 2.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.8 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.2 | 0.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 1.0 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.1 | 2.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 5.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 6.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 1.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 4.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 1.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 1.8 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.6 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.2 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) |
| 0.1 | 0.2 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.0 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 3.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 2.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0033814 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 9.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 4.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 2.4 | GO:0003729 | mRNA binding(GO:0003729) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 5.2 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 7.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |