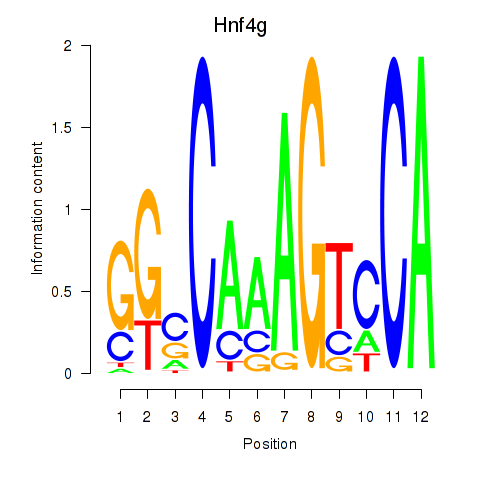
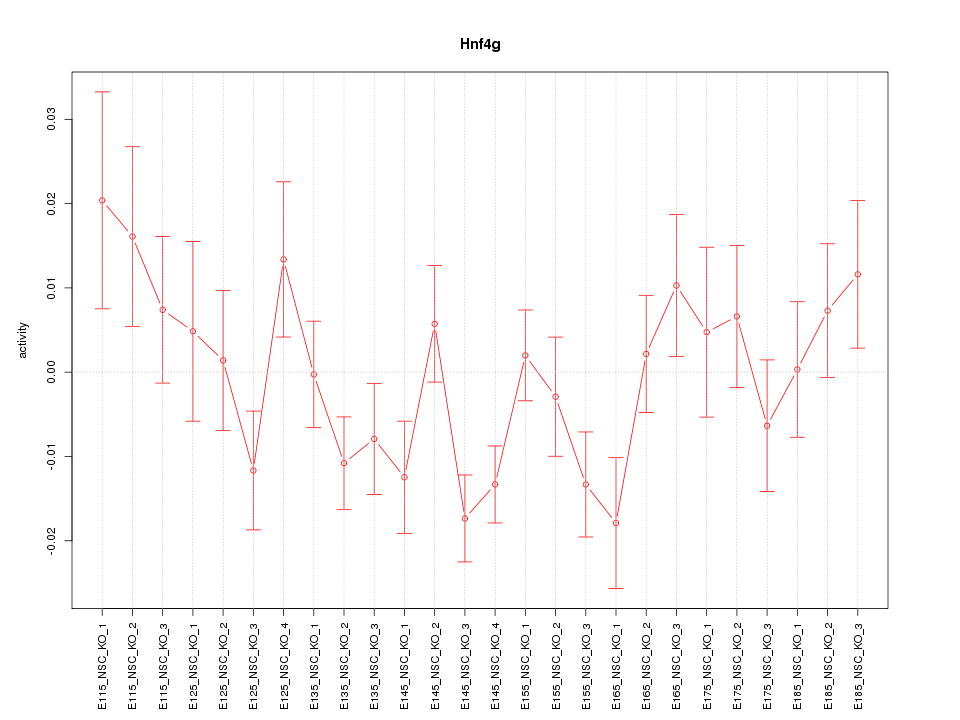
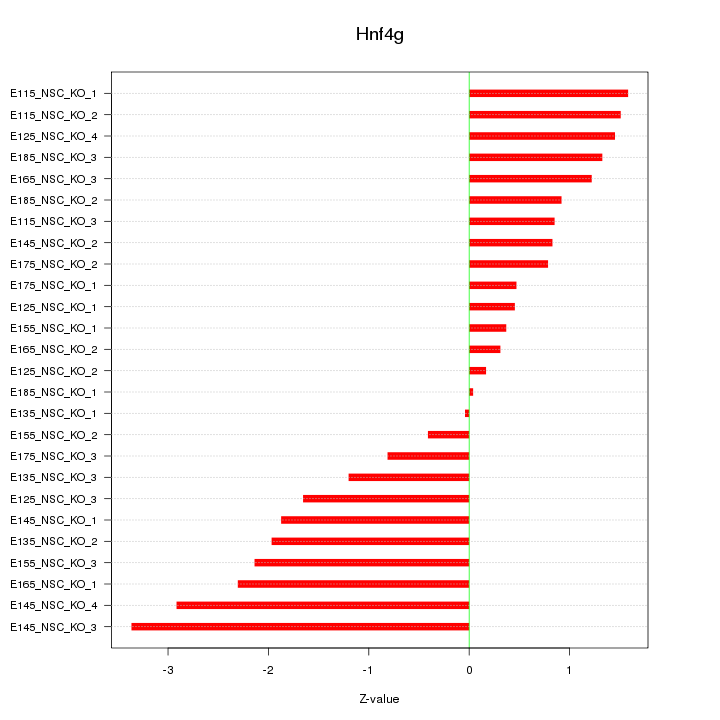
| 0.9 |
2.6 |
GO:0034310 |
primary alcohol catabolic process(GO:0034310) |
| 0.6 |
1.8 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.4 |
1.3 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 |
1.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.4 |
1.6 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.4 |
1.5 |
GO:0070858 |
negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.4 |
1.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.4 |
1.1 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 |
1.0 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.3 |
1.0 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 |
0.3 |
GO:2000977 |
regulation of forebrain neuron differentiation(GO:2000977) |
| 0.3 |
1.3 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.3 |
0.9 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.3 |
0.9 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.3 |
1.8 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 |
1.1 |
GO:0038145 |
macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.3 |
1.3 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.3 |
1.3 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
0.8 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.3 |
0.8 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.3 |
1.3 |
GO:0009313 |
oligosaccharide catabolic process(GO:0009313) |
| 0.3 |
0.5 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.3 |
0.8 |
GO:0002447 |
eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.2 |
1.7 |
GO:0035360 |
positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) |
| 0.2 |
1.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 |
0.9 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.2 |
1.2 |
GO:2000065 |
negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.2 |
0.7 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 |
0.7 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 |
2.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.2 |
1.7 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 |
0.2 |
GO:0045608 |
inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.2 |
1.5 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.8 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 |
0.2 |
GO:0021523 |
somatic motor neuron differentiation(GO:0021523) |
| 0.2 |
0.6 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.2 |
0.8 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 |
1.0 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.2 |
0.8 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.2 |
2.3 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 |
0.8 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.2 |
0.6 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 |
0.6 |
GO:0071649 |
regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.2 |
1.9 |
GO:1904948 |
midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.2 |
0.9 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 |
0.9 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.2 |
0.5 |
GO:0007354 |
zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.2 |
1.0 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 |
1.2 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 |
1.0 |
GO:0010694 |
positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.2 |
0.9 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.2 |
0.5 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.2 |
0.5 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 |
1.8 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 |
0.8 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.2 |
1.5 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.2 |
0.6 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.2 |
0.3 |
GO:0015675 |
nickel cation transport(GO:0015675) |
| 0.2 |
0.5 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 |
1.1 |
GO:0042148 |
strand invasion(GO:0042148) |
| 0.2 |
0.5 |
GO:0046061 |
dATP catabolic process(GO:0046061) |
| 0.2 |
0.5 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.2 |
2.0 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 |
0.6 |
GO:2000256 |
positive regulation of male germ cell proliferation(GO:2000256) |
| 0.1 |
0.7 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.1 |
GO:0071672 |
negative regulation of smooth muscle cell chemotaxis(GO:0071672) |
| 0.1 |
0.7 |
GO:0046654 |
tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 |
0.9 |
GO:0060768 |
regulation of epithelial cell proliferation involved in prostate gland development(GO:0060768) negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 |
0.4 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.1 |
0.4 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.3 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.7 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 |
0.7 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.4 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.9 |
GO:0061620 |
glycolytic process through glucose-6-phosphate(GO:0061620) |
| 0.1 |
0.5 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
0.4 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
0.8 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.1 |
0.4 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 |
1.3 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.1 |
0.4 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.1 |
0.8 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.3 |
GO:0043096 |
purine nucleobase salvage(GO:0043096) |
| 0.1 |
0.4 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.3 |
GO:0071671 |
regulation of smooth muscle cell chemotaxis(GO:0071671) positive regulation of smooth muscle cell chemotaxis(GO:0071673) |
| 0.1 |
0.4 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.6 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.9 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.1 |
1.1 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 |
0.4 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.2 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 |
1.2 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
0.2 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 |
0.4 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.1 |
0.5 |
GO:0010633 |
negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 |
0.4 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 |
0.1 |
GO:0097296 |
activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 |
0.6 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.5 |
GO:0031214 |
biomineral tissue development(GO:0031214) |
| 0.1 |
0.4 |
GO:0090032 |
operant conditioning(GO:0035106) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.1 |
0.4 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.2 |
GO:0097066 |
response to thyroid hormone(GO:0097066) |
| 0.1 |
2.0 |
GO:0070286 |
axonemal dynein complex assembly(GO:0070286) |
| 0.1 |
0.2 |
GO:0060754 |
positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 |
0.6 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 |
0.3 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
0.3 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.2 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.1 |
0.7 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.1 |
1.0 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
0.8 |
GO:0042511 |
positive regulation of tyrosine phosphorylation of Stat1 protein(GO:0042511) |
| 0.1 |
0.1 |
GO:1900020 |
regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 |
0.8 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.3 |
GO:0010752 |
signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.1 |
0.4 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.1 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.1 |
0.3 |
GO:0035811 |
negative regulation of urine volume(GO:0035811) |
| 0.1 |
0.4 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 |
0.6 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.6 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 |
0.6 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.1 |
0.3 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
0.4 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
1.8 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
0.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.7 |
GO:0032725 |
positive regulation of granulocyte macrophage colony-stimulating factor production(GO:0032725) |
| 0.1 |
0.6 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.1 |
0.8 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 |
0.5 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
1.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.1 |
0.2 |
GO:0046618 |
drug export(GO:0046618) |
| 0.1 |
0.1 |
GO:1903020 |
positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 |
0.9 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.1 |
0.5 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.1 |
0.5 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
0.3 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.6 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 |
0.3 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.6 |
GO:0061051 |
positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 |
0.3 |
GO:0014891 |
striated muscle atrophy(GO:0014891) |
| 0.1 |
0.3 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.1 |
5.5 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.1 |
0.2 |
GO:0060216 |
definitive hemopoiesis(GO:0060216) |
| 0.1 |
0.3 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 |
0.2 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.1 |
1.5 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
1.3 |
GO:0060261 |
positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.1 |
2.3 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.1 |
0.2 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.2 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 |
0.2 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.3 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 |
0.2 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 |
0.1 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.4 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
0.5 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.1 |
0.3 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 |
0.1 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.1 |
0.6 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
0.7 |
GO:0015695 |
organic cation transport(GO:0015695) |
| 0.1 |
0.4 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
0.1 |
GO:0051953 |
negative regulation of organic acid transport(GO:0032891) negative regulation of amine transport(GO:0051953) negative regulation of amino acid transport(GO:0051956) |
| 0.1 |
0.1 |
GO:0043585 |
nose morphogenesis(GO:0043585) |
| 0.1 |
0.3 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.1 |
0.3 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.1 |
0.7 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 |
0.5 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.2 |
GO:0016068 |
regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.1 |
0.7 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.3 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.2 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.9 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 |
0.6 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.1 |
0.2 |
GO:0046544 |
development of secondary male sexual characteristics(GO:0046544) |
| 0.1 |
0.3 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.1 |
0.3 |
GO:0051006 |
positive regulation of lipoprotein lipase activity(GO:0051006) |
| 0.1 |
0.2 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.6 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.1 |
0.3 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.1 |
0.4 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
0.5 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.1 |
0.7 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.1 |
0.2 |
GO:2000097 |
regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 |
0.4 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
0.4 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
0.2 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.1 |
0.2 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
0.8 |
GO:0060055 |
angiogenesis involved in wound healing(GO:0060055) |
| 0.1 |
0.4 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
0.1 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.1 |
0.2 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
0.5 |
GO:0051958 |
methotrexate transport(GO:0051958) |
| 0.1 |
0.1 |
GO:0060155 |
platelet dense granule organization(GO:0060155) |
| 0.1 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.6 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 |
2.4 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.2 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 |
0.9 |
GO:0009437 |
carnitine metabolic process(GO:0009437) |
| 0.1 |
0.2 |
GO:0006449 |
regulation of translational termination(GO:0006449) |
| 0.1 |
0.1 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.2 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
0.3 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 |
0.3 |
GO:0072383 |
plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 |
0.5 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.6 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.1 |
0.1 |
GO:1990705 |
cholangiocyte proliferation(GO:1990705) |
| 0.1 |
0.4 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
0.2 |
GO:0060120 |
auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 |
0.2 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.1 |
0.5 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.1 |
0.2 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.1 |
0.7 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.1 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.1 |
0.3 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) |
| 0.1 |
0.5 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.1 |
0.5 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.1 |
GO:1901228 |
positive regulation of skeletal muscle tissue growth(GO:0048633) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.1 |
0.3 |
GO:0042538 |
hyperosmotic salinity response(GO:0042538) |
| 0.1 |
0.3 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 |
0.3 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 |
0.3 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.1 |
0.7 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.1 |
0.3 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.1 |
GO:0008078 |
mesodermal cell migration(GO:0008078) |
| 0.1 |
0.4 |
GO:0071474 |
cellular hyperosmotic response(GO:0071474) |
| 0.1 |
0.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 |
0.4 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.4 |
GO:0043173 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) nucleotide salvage(GO:0043173) |
| 0.1 |
1.4 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
0.4 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.6 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.1 |
0.4 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 |
0.7 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.1 |
0.3 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.1 |
1.4 |
GO:0031290 |
retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 |
0.3 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 |
0.2 |
GO:0060594 |
mammary gland specification(GO:0060594) |
| 0.1 |
0.1 |
GO:1900864 |
mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 |
1.4 |
GO:0008299 |
isoprenoid biosynthetic process(GO:0008299) |
| 0.1 |
0.1 |
GO:0003057 |
regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.1 |
0.1 |
GO:0006925 |
inflammatory cell apoptotic process(GO:0006925) |
| 0.1 |
0.5 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.2 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.1 |
0.8 |
GO:0007213 |
G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 |
0.3 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.3 |
GO:0055089 |
fatty acid homeostasis(GO:0055089) |
| 0.1 |
0.2 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.5 |
GO:0030220 |
platelet formation(GO:0030220) |
| 0.1 |
0.2 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.1 |
0.3 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
0.2 |
GO:0051354 |
negative regulation of oxidoreductase activity(GO:0051354) |
| 0.1 |
0.2 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.1 |
0.2 |
GO:0033313 |
meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 |
0.1 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.4 |
GO:0006004 |
fucose metabolic process(GO:0006004) |
| 0.1 |
0.3 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
0.2 |
GO:0009063 |
cellular amino acid catabolic process(GO:0009063) |
| 0.1 |
0.2 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
0.3 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 |
0.1 |
GO:0046078 |
pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) dUMP metabolic process(GO:0046078) |
| 0.1 |
1.3 |
GO:0003416 |
endochondral bone growth(GO:0003416) |
| 0.1 |
0.4 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 |
0.2 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 |
0.3 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 |
0.4 |
GO:1902166 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 |
0.2 |
GO:0002086 |
diaphragm contraction(GO:0002086) |
| 0.1 |
0.5 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.1 |
0.2 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.1 |
0.3 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
0.2 |
GO:0032687 |
negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 |
0.2 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.1 |
0.2 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.5 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.1 |
0.3 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.3 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.5 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.3 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.2 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 |
0.2 |
GO:0061036 |
positive regulation of cartilage development(GO:0061036) |
| 0.1 |
0.2 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 |
0.2 |
GO:0015819 |
lysine transport(GO:0015819) |
| 0.1 |
0.2 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 |
0.6 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.2 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 |
0.1 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 |
0.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.3 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.3 |
GO:0006573 |
valine metabolic process(GO:0006573) |
| 0.0 |
0.9 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.0 |
1.6 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.0 |
0.3 |
GO:0042135 |
neurotransmitter catabolic process(GO:0042135) |
| 0.0 |
0.4 |
GO:0070307 |
lens fiber cell development(GO:0070307) |
| 0.0 |
0.2 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 |
0.3 |
GO:0046459 |
short-chain fatty acid metabolic process(GO:0046459) |
| 0.0 |
1.4 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.1 |
GO:0006533 |
aspartate catabolic process(GO:0006533) |
| 0.0 |
0.3 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.2 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.0 |
0.2 |
GO:0051001 |
negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 |
0.2 |
GO:0035511 |
oxidative DNA demethylation(GO:0035511) |
| 0.0 |
0.1 |
GO:0060982 |
coronary artery morphogenesis(GO:0060982) |
| 0.0 |
0.6 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
1.5 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.6 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.2 |
GO:0044828 |
negative regulation by host of viral process(GO:0044793) negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 |
0.3 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
1.0 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.1 |
GO:0090237 |
regulation of arachidonic acid secretion(GO:0090237) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 |
0.5 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.3 |
GO:0006528 |
asparagine metabolic process(GO:0006528) |
| 0.0 |
0.2 |
GO:0032429 |
regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 |
0.3 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.5 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.9 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.6 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.6 |
GO:0042493 |
response to drug(GO:0042493) |
| 0.0 |
1.0 |
GO:0001937 |
negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 |
0.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
1.6 |
GO:0043297 |
apical junction assembly(GO:0043297) |
| 0.0 |
0.1 |
GO:0003192 |
mitral valve formation(GO:0003192) cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 |
0.3 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.0 |
0.2 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 |
0.2 |
GO:0046825 |
regulation of protein export from nucleus(GO:0046825) |
| 0.0 |
0.5 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.1 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.1 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 |
0.2 |
GO:0006207 |
'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 |
0.2 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.0 |
0.3 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.1 |
GO:0035793 |
negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:0042362 |
fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 |
0.1 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.0 |
0.1 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.0 |
0.7 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.2 |
GO:2001198 |
regulation of dendritic cell differentiation(GO:2001198) negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 |
0.3 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.1 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.1 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.4 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.9 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
2.2 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
0.1 |
GO:0048755 |
branching morphogenesis of a nerve(GO:0048755) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.4 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
0.4 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.0 |
0.9 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.9 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.1 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
0.1 |
GO:0061101 |
neuroendocrine cell differentiation(GO:0061101) |
| 0.0 |
0.6 |
GO:0090022 |
regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 |
0.2 |
GO:0032074 |
negative regulation of nuclease activity(GO:0032074) |
| 0.0 |
0.8 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.2 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.3 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.7 |
GO:0006631 |
fatty acid metabolic process(GO:0006631) |
| 0.0 |
0.1 |
GO:0050812 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 |
0.5 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.4 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 |
0.1 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 |
0.1 |
GO:0045002 |
double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.0 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 |
0.9 |
GO:0035136 |
forelimb morphogenesis(GO:0035136) |
| 0.0 |
0.1 |
GO:0060398 |
regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 |
0.2 |
GO:0031018 |
endocrine pancreas development(GO:0031018) |
| 0.0 |
0.7 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.2 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 |
0.4 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.4 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.0 |
0.3 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 |
0.0 |
GO:0032364 |
oxygen homeostasis(GO:0032364) |
| 0.0 |
0.1 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 |
0.2 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.4 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 |
0.3 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 |
0.1 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.2 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.5 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.1 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 |
0.1 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 |
0.3 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.1 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.1 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) aromatic amino acid family catabolic process(GO:0009074) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.0 |
1.3 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.2 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.1 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 |
0.2 |
GO:0033033 |
negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 |
0.1 |
GO:0002281 |
macrophage activation involved in immune response(GO:0002281) |
| 0.0 |
0.2 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.0 |
0.2 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 |
0.1 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.3 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.1 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.0 |
0.2 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 |
0.3 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.5 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.8 |
GO:0007584 |
response to nutrient(GO:0007584) |
| 0.0 |
0.3 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.5 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 |
0.2 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.4 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.9 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
0.1 |
GO:0048563 |
post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 |
0.3 |
GO:0048643 |
positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 |
0.2 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.1 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
0.4 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.2 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 |
0.1 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:0030578 |
PML body organization(GO:0030578) |
| 0.0 |
0.1 |
GO:1902254 |
regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:1901550 |
regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 |
0.2 |
GO:0045333 |
cellular respiration(GO:0045333) |
| 0.0 |
0.1 |
GO:0072061 |
inner medullary collecting duct development(GO:0072061) |
| 0.0 |
0.1 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.4 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.1 |
GO:0040032 |
post-embryonic body morphogenesis(GO:0040032) |
| 0.0 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.0 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 |
0.1 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.2 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.0 |
0.1 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.6 |
GO:0046782 |
regulation of viral transcription(GO:0046782) |
| 0.0 |
0.1 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.1 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.0 |
GO:0060426 |
lung vasculature development(GO:0060426) |
| 0.0 |
0.5 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.2 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
0.1 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.0 |
0.5 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.3 |
GO:0045880 |
positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 |
0.1 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.1 |
GO:0043129 |
surfactant homeostasis(GO:0043129) |
| 0.0 |
0.0 |
GO:0006103 |
2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 |
0.6 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.9 |
GO:0002062 |
chondrocyte differentiation(GO:0002062) |
| 0.0 |
0.8 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.6 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.0 |
0.2 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
0.0 |
GO:0061724 |
lipophagy(GO:0061724) |
| 0.0 |
0.4 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.0 |
0.2 |
GO:0035269 |
protein O-linked mannosylation(GO:0035269) |
| 0.0 |
0.1 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.1 |
GO:0031666 |
positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 |
0.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 |
0.5 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.6 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.3 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.1 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.0 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
0.6 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.6 |
GO:0042168 |
heme metabolic process(GO:0042168) |
| 0.0 |
0.5 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.1 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
1.1 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.0 |
0.6 |
GO:0002066 |
columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 |
0.1 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.0 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) rRNA transport(GO:0051029) |
| 0.0 |
0.0 |
GO:1901838 |
positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 |
0.1 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.2 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0042517 |
positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.0 |
0.2 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.1 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.0 |
0.0 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.5 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.1 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.1 |
GO:0072602 |
interleukin-4 secretion(GO:0072602) |
| 0.0 |
0.1 |
GO:0014068 |
positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 |
0.1 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.4 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.5 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.1 |
GO:0042990 |
regulation of transcription factor import into nucleus(GO:0042990) |
| 0.0 |
0.0 |
GO:0090241 |
negative regulation of histone H4 acetylation(GO:0090241) |
| 0.0 |
0.3 |
GO:0050673 |
epithelial cell proliferation(GO:0050673) |
| 0.0 |
0.3 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.0 |
0.1 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.1 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.0 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 |
0.1 |
GO:0032776 |
DNA methylation on cytosine within a CG sequence(GO:0010424) DNA methylation on cytosine(GO:0032776) |
| 0.0 |
0.1 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.3 |
GO:0006582 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.8 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.0 |
0.1 |
GO:0006309 |
apoptotic DNA fragmentation(GO:0006309) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.1 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.2 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.0 |
GO:0046166 |
glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 |
0.4 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.1 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.1 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.1 |
GO:0009081 |
branched-chain amino acid metabolic process(GO:0009081) branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.0 |
GO:0072393 |
microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 |
0.1 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.1 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
0.1 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.0 |
0.5 |
GO:0032648 |
regulation of interferon-beta production(GO:0032648) |
| 0.0 |
0.3 |
GO:0016575 |
histone deacetylation(GO:0016575) |
| 0.0 |
0.1 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.1 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.0 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.0 |
0.1 |
GO:2000191 |
regulation of fatty acid transport(GO:2000191) |
| 0.0 |
0.1 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.1 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 |
0.1 |
GO:0046622 |
positive regulation of organ growth(GO:0046622) |
| 0.0 |
0.3 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.0 |
GO:0034035 |
sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 |
0.2 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.0 |
GO:1904152 |
regulation of retrograde protein transport, ER to cytosol(GO:1904152) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.0 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.1 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.0 |
0.1 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.3 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
0.0 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.0 |
GO:0032466 |
negative regulation of cytokinesis(GO:0032466) |
| 0.0 |
0.1 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.0 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.0 |
GO:0038089 |
positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) protein kinase D signaling(GO:0089700) |
| 0.0 |
0.1 |
GO:0097205 |
renal filtration(GO:0097205) |
| 0.0 |
0.1 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |