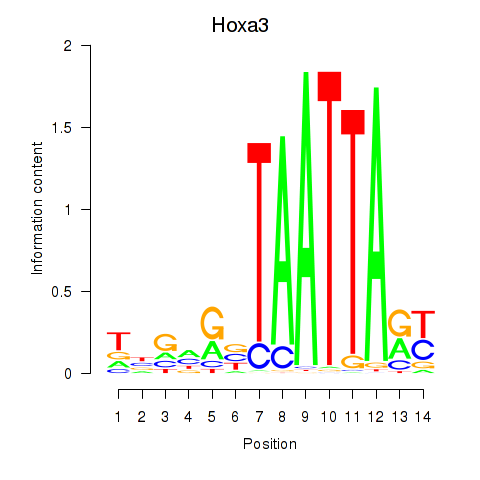
| 0.5 |
1.6 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.2 |
3.8 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.2 |
0.5 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
2.3 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
1.5 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
1.5 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.0 |
1.9 |
GO:0051693 |
actin filament capping(GO:0051693) |
| 0.0 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.2 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.6 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |