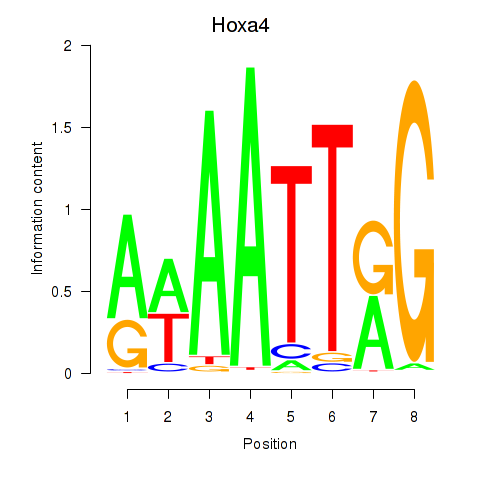
| 1.6 |
4.8 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.3 |
3.9 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 1.2 |
3.5 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 1.0 |
2.9 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.8 |
8.9 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.7 |
3.0 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.4 |
1.3 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.4 |
1.1 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.3 |
2.4 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.3 |
1.0 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.3 |
3.4 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.3 |
0.6 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.3 |
2.5 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 |
0.8 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.3 |
1.3 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.3 |
2.8 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.3 |
2.0 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 |
0.8 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.2 |
0.9 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.2 |
1.9 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.2 |
6.6 |
GO:0045838 |
positive regulation of membrane potential(GO:0045838) |
| 0.2 |
1.5 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 |
1.0 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.2 |
1.8 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
0.5 |
GO:0003032 |
detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.2 |
1.0 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.2 |
0.6 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
0.4 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 |
0.5 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.8 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.1 |
0.7 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.4 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 |
1.5 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
1.0 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.7 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
7.4 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
0.8 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
1.0 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
4.6 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.1 |
1.0 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.9 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.1 |
0.7 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.1 |
0.8 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 |
1.4 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.1 |
1.6 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.1 |
1.2 |
GO:0061050 |
regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 |
0.7 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
1.0 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.1 |
0.4 |
GO:0042574 |
isoprenoid catabolic process(GO:0008300) retinal metabolic process(GO:0042574) |
| 0.0 |
0.8 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.2 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.2 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 |
0.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.9 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.8 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.5 |
GO:0044144 |
regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 |
1.3 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
3.1 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.7 |
GO:0016556 |
mRNA modification(GO:0016556) |
| 0.0 |
0.2 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 |
1.0 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
2.2 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.5 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 |
0.3 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.6 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
1.2 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
2.0 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
1.0 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
1.7 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.3 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
1.5 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.1 |
GO:0051775 |
response to redox state(GO:0051775) |
| 0.0 |
2.0 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.4 |
GO:0034389 |
lipid particle organization(GO:0034389) negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 |
0.4 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.1 |
GO:2001013 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 |
1.8 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.2 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.0 |
0.5 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.6 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
2.3 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.3 |
GO:0071480 |
cellular response to gamma radiation(GO:0071480) |
| 0.0 |
0.6 |
GO:2000179 |
positive regulation of neural precursor cell proliferation(GO:2000179) |