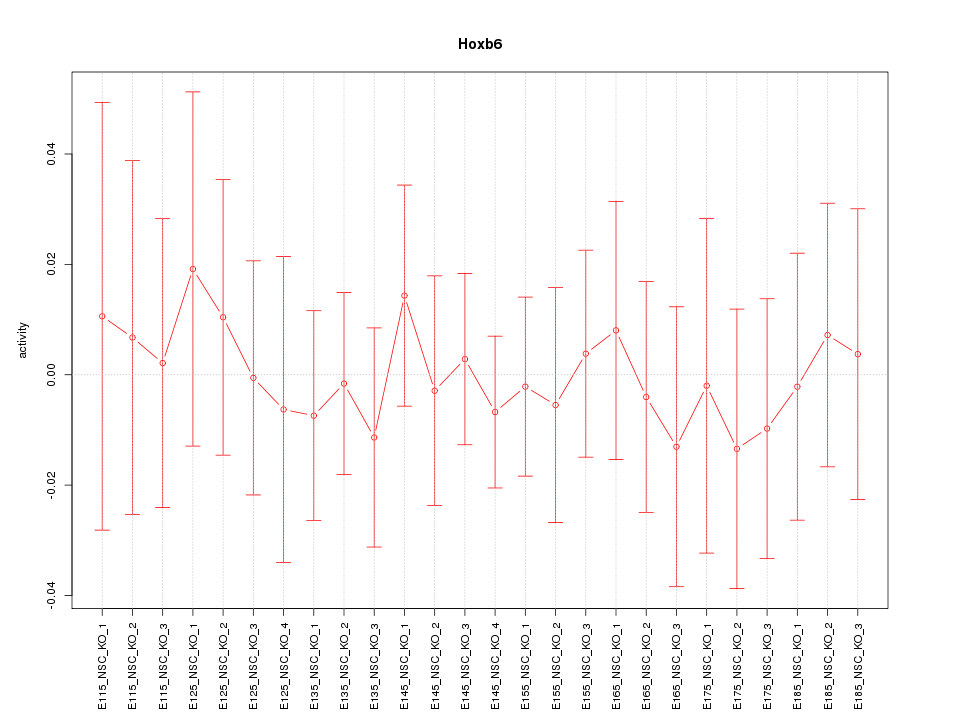
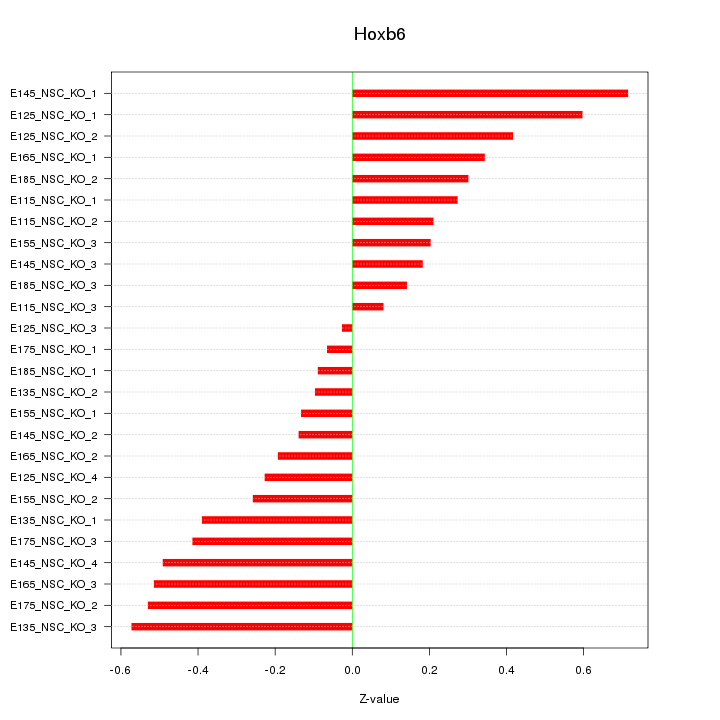
| 0.2 |
1.1 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 |
0.5 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 |
0.3 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 |
0.4 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.1 |
0.6 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.5 |
GO:0021559 |
trigeminal nerve development(GO:0021559) |
| 0.0 |
0.8 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.9 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.2 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.1 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) regulation of microtubule motor activity(GO:2000574) |
| 0.0 |
0.4 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.1 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 |
0.2 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.2 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.0 |
0.1 |
GO:0006012 |
galactose metabolic process(GO:0006012) |