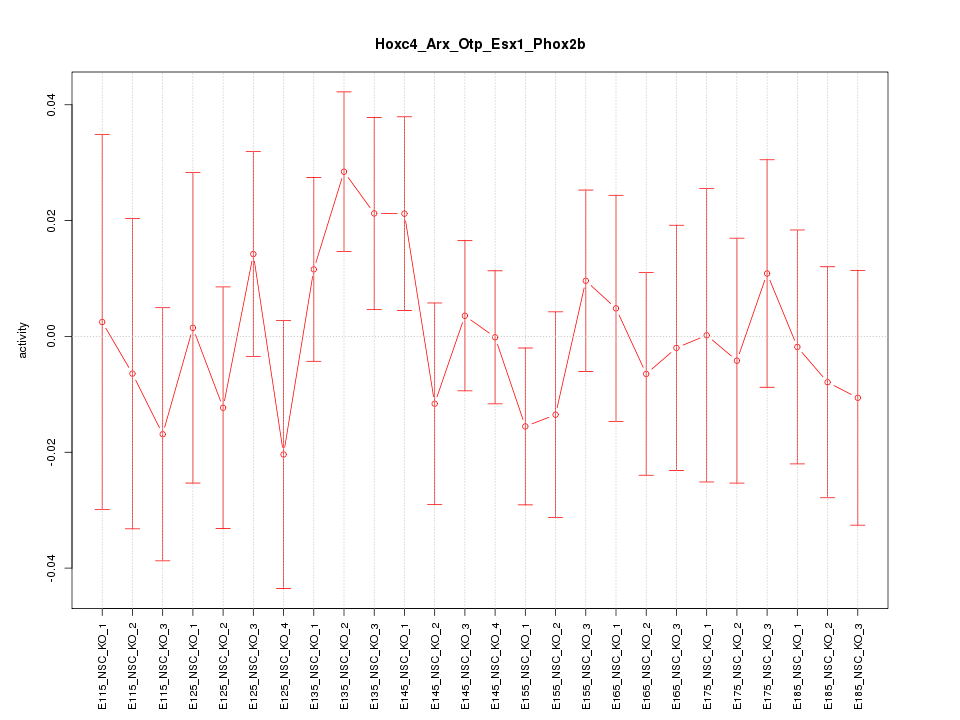
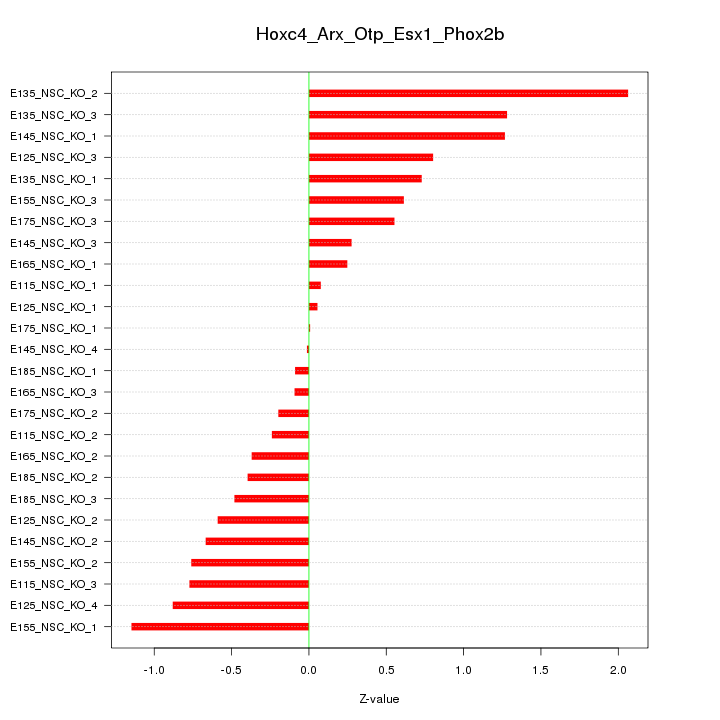
Motif ID: Hoxc4_Arx_Otp_Esx1_Phox2b
Z-value: 0.739
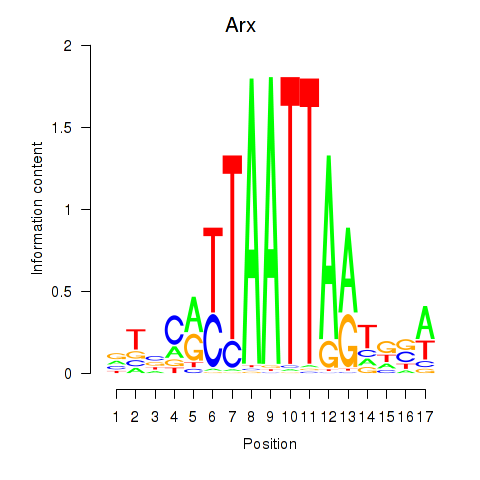
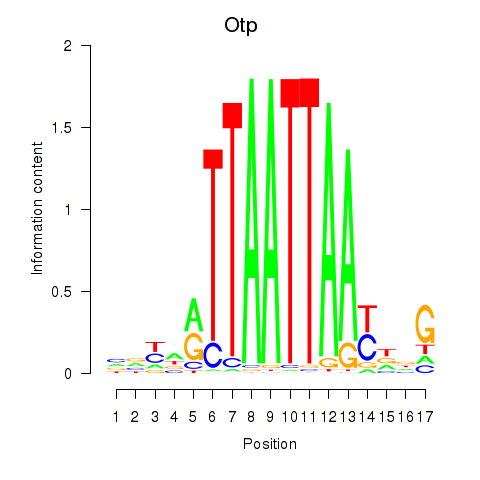
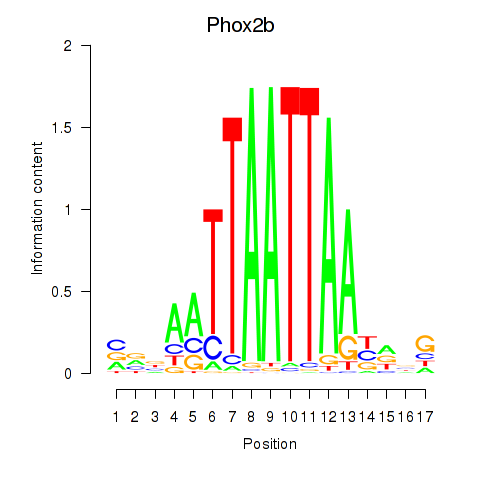
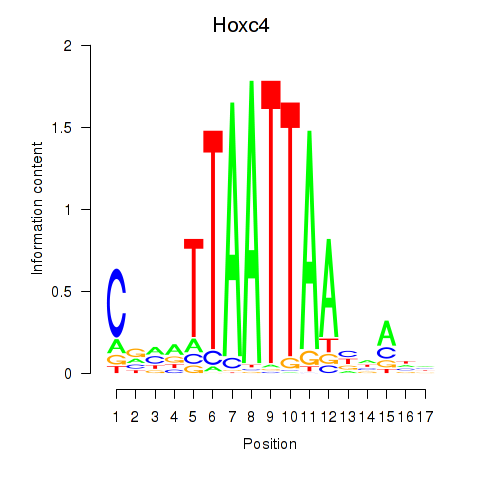
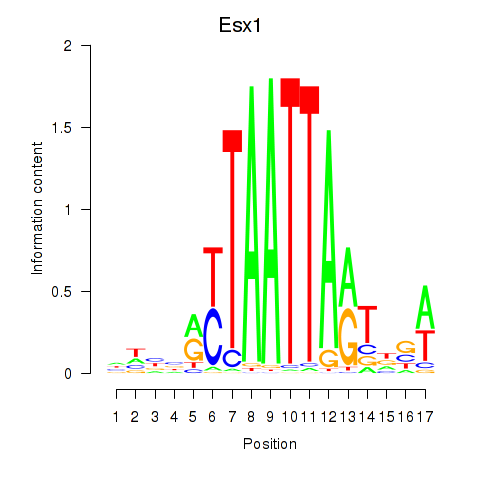
Transcription factors associated with Hoxc4_Arx_Otp_Esx1_Phox2b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Arx | ENSMUSG00000035277.9 | Arx |
| Esx1 | ENSMUSG00000023443.7 | Esx1 |
| Hoxc4 | ENSMUSG00000075394.3 | Hoxc4 |
| Otp | ENSMUSG00000021685.10 | Otp |
| Phox2b | ENSMUSG00000012520.8 | Phox2b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Arx | mm10_v2_chrX_+_93286499_93286522 | 0.19 | 3.5e-01 | Click! |
| Otp | mm10_v2_chr13_+_94875600_94875611 | -0.10 | 6.2e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.0 | GO:0021856 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 | 0.2 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) oxidative demethylation(GO:0070989) |
| 0.2 | 0.5 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.5 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 1.4 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.2 | GO:0043987 | histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.1 | GO:0046098 | regulation of primitive erythrocyte differentiation(GO:0010725) guanine metabolic process(GO:0046098) |
| 0.0 | 0.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0072592 | regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.3 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.0 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.3 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.0 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0051378 | primary amine oxidase activity(GO:0008131) serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.4 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |