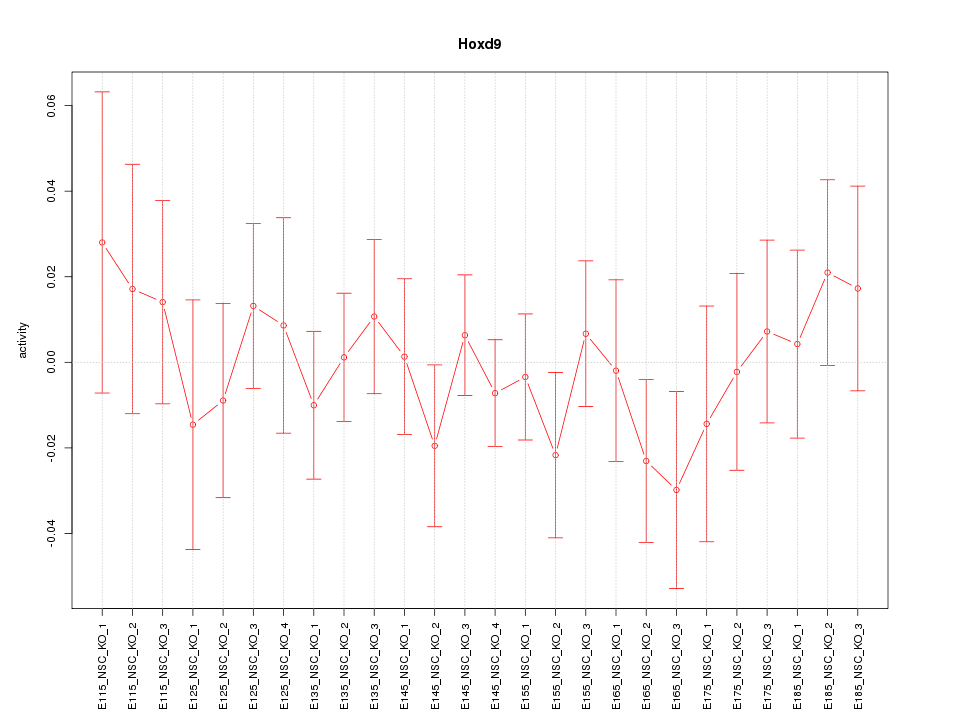
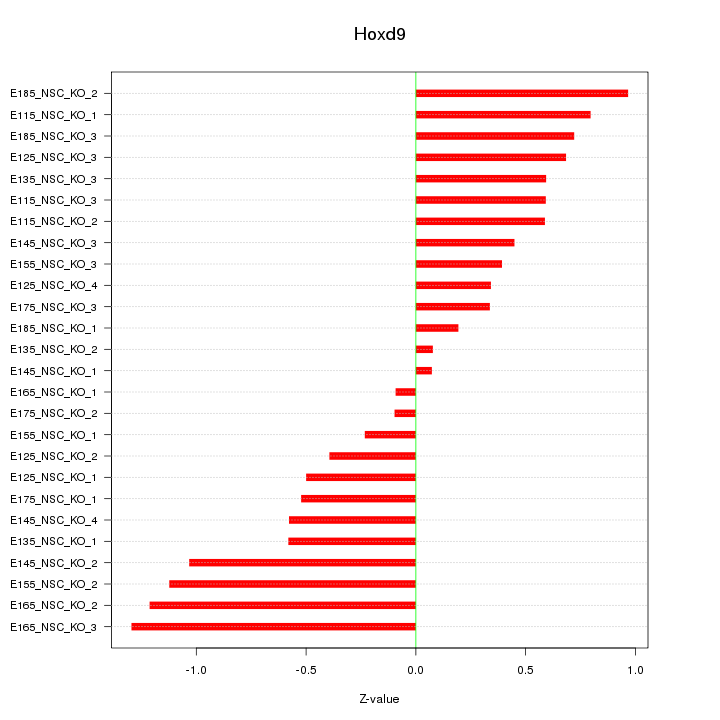
Motif ID: Hoxd9
Z-value: 0.654
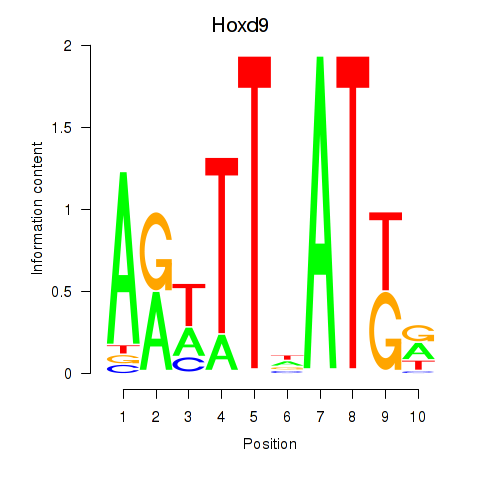
Transcription factors associated with Hoxd9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd9 | ENSMUSG00000043342.8 | Hoxd9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 | 1.7 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.5 | 2.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 0.8 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 2.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.4 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.3 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.1 | 0.3 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 2.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.8 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.7 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.2 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.0 | 0.3 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 | 0.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.4 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.9 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 1.0 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.4 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.3 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 2.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.0 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.0 | 0.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.0 | 0.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 2.8 | GO:0017166 | dystroglycan binding(GO:0002162) vinculin binding(GO:0017166) |
| 0.2 | 1.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.5 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 2.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |