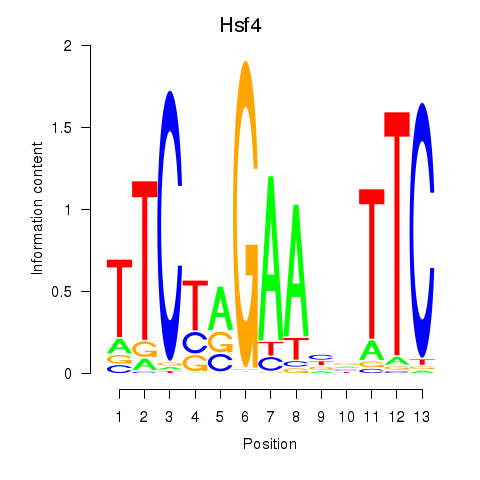
| 0.6 |
3.5 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.3 |
1.0 |
GO:0035799 |
ureter maturation(GO:0035799) |
| 0.2 |
0.2 |
GO:0061113 |
pancreas morphogenesis(GO:0061113) |
| 0.2 |
0.5 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
0.7 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.6 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.5 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 |
1.0 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.3 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 |
0.4 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.1 |
0.3 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 |
0.4 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 |
0.3 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.5 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.3 |
GO:1900041 |
negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 |
0.3 |
GO:0019043 |
establishment of viral latency(GO:0019043) |
| 0.1 |
0.4 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.1 |
0.3 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 |
0.2 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.2 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.1 |
0.3 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
0.4 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 |
0.4 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.2 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 0.1 |
0.2 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.3 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.1 |
0.3 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 |
0.5 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 |
0.5 |
GO:1900107 |
regulation of nodal signaling pathway(GO:1900107) |
| 0.1 |
0.3 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
0.4 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
0.2 |
GO:1904154 |
protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.2 |
GO:0048698 |
negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 |
0.4 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.2 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.2 |
GO:0010248 |
B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.1 |
0.8 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 0.1 |
0.6 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.3 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 |
0.3 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.4 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.2 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.1 |
0.5 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.3 |
GO:0002326 |
B cell lineage commitment(GO:0002326) |
| 0.0 |
0.3 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 |
1.6 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.5 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.2 |
GO:1901163 |
trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) |
| 0.0 |
0.3 |
GO:0006590 |
thyroid hormone generation(GO:0006590) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.5 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.4 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.2 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0031117 |
serotonin receptor signaling pathway(GO:0007210) positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.2 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 |
0.1 |
GO:1903054 |
lysosomal membrane organization(GO:0097212) negative regulation of extracellular matrix organization(GO:1903054) positive regulation of protein folding(GO:1903334) |
| 0.0 |
0.1 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.0 |
0.7 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.4 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.0 |
0.4 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.3 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.1 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 |
0.4 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.2 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.1 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.2 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.0 |
0.1 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 |
0.1 |
GO:0035701 |
hematopoietic stem cell migration(GO:0035701) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.2 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 |
0.3 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.2 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.1 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.1 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.3 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.2 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.0 |
0.9 |
GO:0021591 |
ventricular system development(GO:0021591) |
| 0.0 |
0.2 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.1 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 |
0.2 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.0 |
0.2 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.2 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.0 |
0.2 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.0 |
0.1 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 |
0.0 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.1 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:0097460 |
ferrous iron import into cell(GO:0097460) |
| 0.0 |
0.2 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.1 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.2 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.1 |
GO:0032926 |
negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 |
0.1 |
GO:0051036 |
regulation of endosome size(GO:0051036) receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.4 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) negative regulation of sprouting angiogenesis(GO:1903671) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.3 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.1 |
GO:0090037 |
positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 |
0.3 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.2 |
GO:0003094 |
glomerular filtration(GO:0003094) |
| 0.0 |
0.2 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.1 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.0 |
0.5 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.0 |
GO:0002924 |
positive regulation of tolerance induction(GO:0002645) negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 |
0.1 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.1 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |