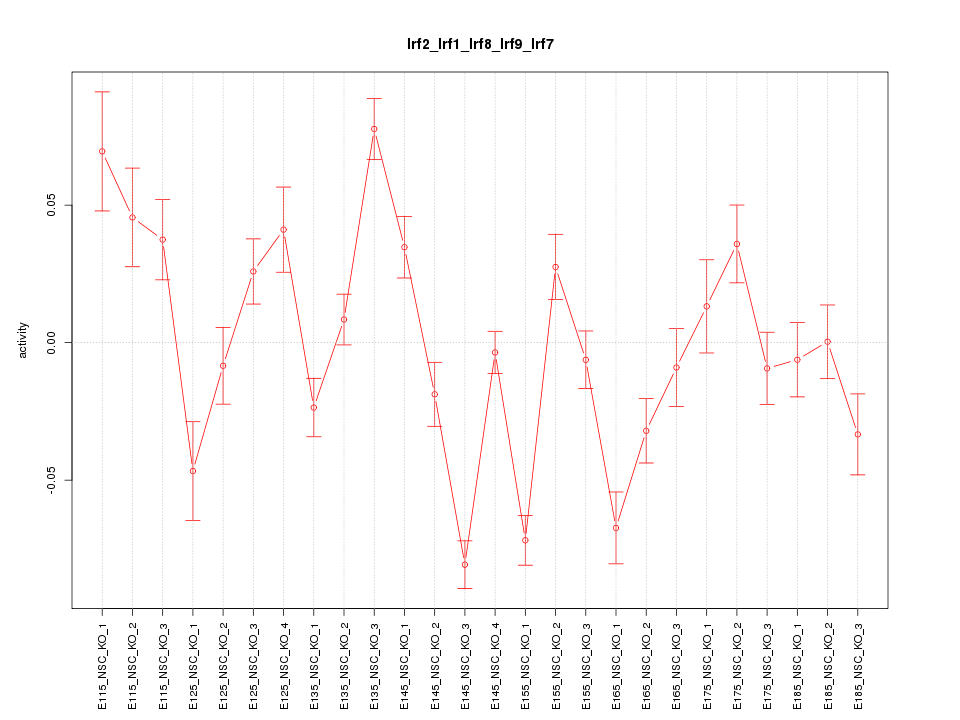
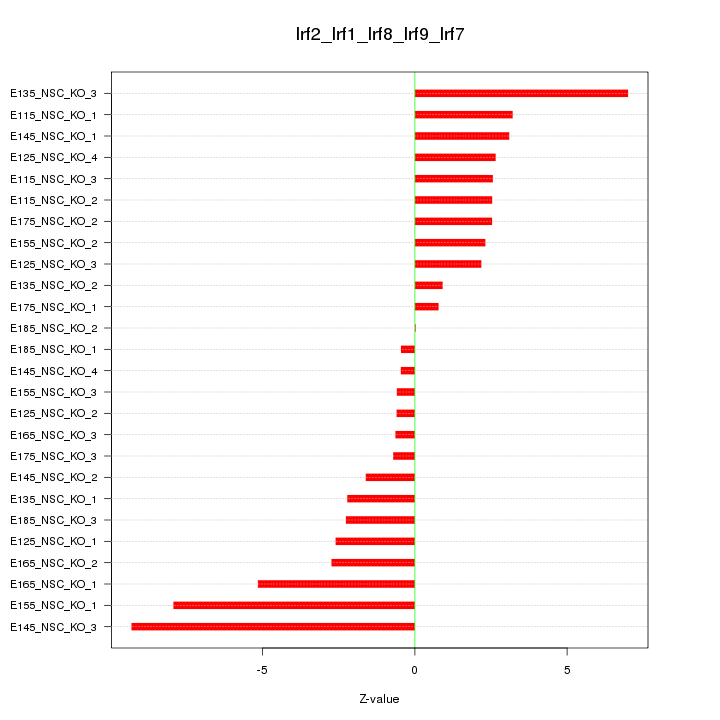
Motif ID: Irf2_Irf1_Irf8_Irf9_Irf7
Z-value: 3.465
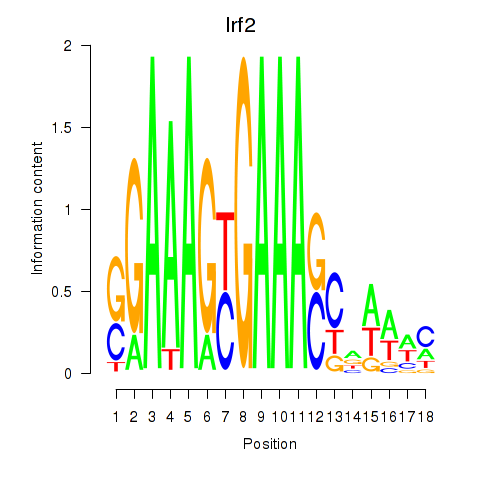
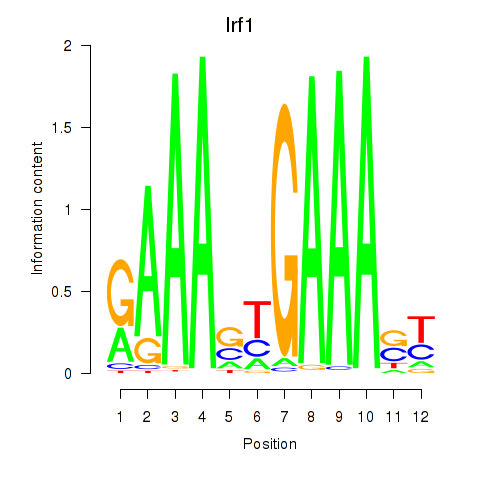
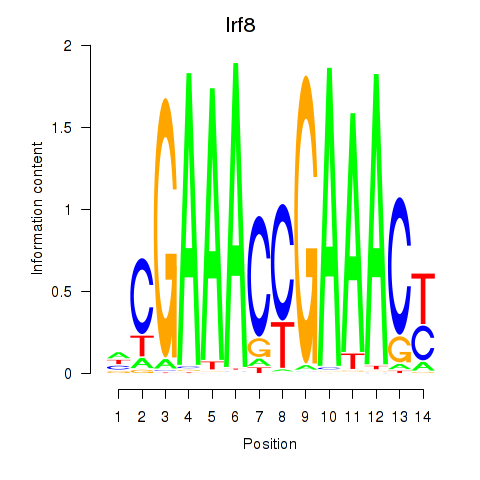
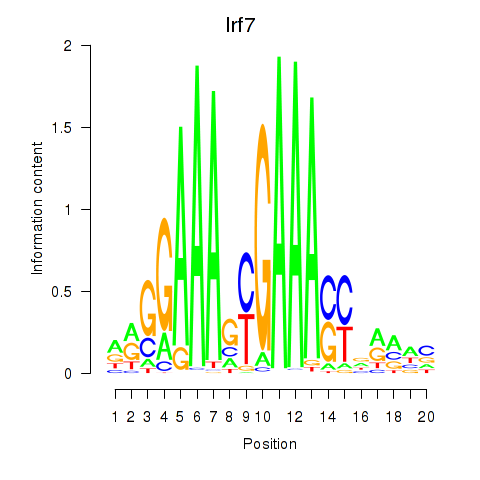
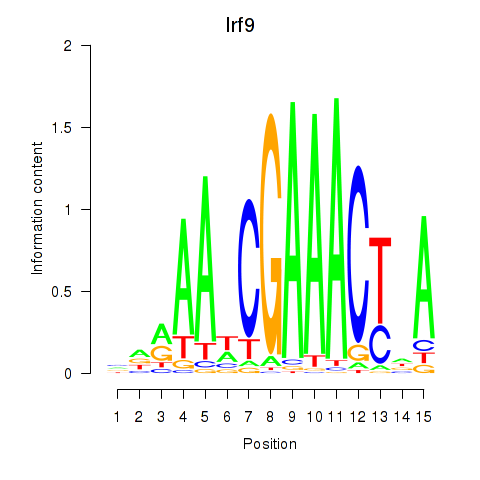
Transcription factors associated with Irf2_Irf1_Irf8_Irf9_Irf7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Irf1 | ENSMUSG00000018899.10 | Irf1 |
| Irf2 | ENSMUSG00000031627.7 | Irf2 |
| Irf7 | ENSMUSG00000025498.8 | Irf7 |
| Irf8 | ENSMUSG00000041515.3 | Irf8 |
| Irf9 | ENSMUSG00000002325.8 | Irf9 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf1 | mm10_v2_chr11_+_53770014_53770057 | 0.59 | 1.4e-03 | Click! |
| Irf9 | mm10_v2_chr14_+_55604550_55604579 | 0.41 | 3.9e-02 | Click! |
| Irf7 | mm10_v2_chr7_-_141266415_141266481 | 0.39 | 5.2e-02 | Click! |
| Irf8 | mm10_v2_chr8_+_120736352_120736385 | 0.25 | 2.2e-01 | Click! |
| Irf2 | mm10_v2_chr8_+_46739745_46739791 | 0.08 | 6.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.7 | 8.3 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 1.5 | 7.7 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 1.4 | 5.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.3 | 9.4 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 1.2 | 8.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.1 | 9.0 | GO:1900225 | NLRP3 inflammasome complex assembly(GO:0044546) regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 1.1 | 8.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 1.1 | 3.2 | GO:0032701 | negative regulation of interleukin-18 production(GO:0032701) |
| 1.1 | 3.2 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 1.0 | 2.9 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) quinolinate biosynthetic process(GO:0019805) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 1.0 | 3.8 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.9 | 2.7 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.9 | 2.6 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.8 | 4.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.8 | 2.5 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.8 | 2.3 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 0.8 | 2.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.7 | 2.2 | GO:0002765 | immune response-inhibiting signal transduction(GO:0002765) |
| 0.7 | 2.9 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.7 | 2.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.7 | 2.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 | 1.9 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.6 | 2.5 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 | 1.9 | GO:1990481 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.6 | 2.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.6 | 1.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.5 | 1.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 6.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.5 | 4.8 | GO:0085020 | protein K6-linked ubiquitination(GO:0085020) |
| 0.5 | 1.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.5 | 2.0 | GO:0071316 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of calcidiol 1-monooxygenase activity(GO:0060558) cellular response to nicotine(GO:0071316) response to glycoside(GO:1903416) |
| 0.5 | 3.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.5 | 2.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 1.9 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 1.4 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.4 | 3.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.4 | 2.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.4 | 3.5 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.4 | 2.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.4 | 4.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.4 | 3.0 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 | 1.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 1.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.4 | 1.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.4 | 2.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 2.5 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.3 | 2.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.3 | 0.7 | GO:1900426 | positive regulation of defense response to bacterium(GO:1900426) |
| 0.3 | 2.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 1.0 | GO:0070172 | oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.3 | 1.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.3 | 1.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.3 | 1.3 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.3 | 3.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.3 | 0.9 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.3 | 2.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 4.1 | GO:0097066 | response to thyroid hormone(GO:0097066) |
| 0.3 | 1.2 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.3 | 2.0 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.3 | 0.9 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.3 | 1.7 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.3 | 0.8 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.3 | 0.8 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.3 | 4.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 0.8 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.3 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 3.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 0.8 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.3 | 10.9 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.2 | 1.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.2 | 1.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 1.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.2 | 1.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 0.7 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.2 | 2.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 4.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 0.9 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 1.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.5 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 2.6 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.2 | 1.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 7.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.2 | 2.9 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.2 | 1.0 | GO:0015867 | ATP transport(GO:0015867) |
| 0.2 | 0.8 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.2 | 2.5 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.2 | 1.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 0.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 2.7 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.2 | 3.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.2 | 1.6 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.2 | 3.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 0.7 | GO:1903463 | mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 0.2 | 1.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.7 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 0.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.2 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 0.6 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 0.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.6 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 0.2 | 0.5 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 1.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 0.6 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 0.6 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 0.3 | GO:0097035 | regulation of membrane lipid distribution(GO:0097035) |
| 0.2 | 1.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.2 | 0.9 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 0.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.2 | 0.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 1.3 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.7 | GO:0032459 | regulation of protein oligomerization(GO:0032459) |
| 0.1 | 1.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.3 | GO:0046851 | negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 | 1.9 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.1 | 3.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.3 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.1 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.8 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.1 | 0.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.3 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.1 | 2.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:1904749 | regulation of telomeric DNA binding(GO:1904742) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 1.6 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.1 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.3 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 0.4 | GO:0072592 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) oxygen metabolic process(GO:0072592) positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.6 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.4 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.6 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.6 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 0.9 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 1.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.5 | GO:0046654 | 'de novo' IMP biosynthetic process(GO:0006189) tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.7 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.6 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.1 | 0.2 | GO:1902031 | regulation of pentose-phosphate shunt(GO:0043456) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 3.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 1.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.6 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.3 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.2 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.6 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.1 | 1.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 2.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 2.0 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.3 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.2 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 1.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 3.3 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) paranodal junction assembly(GO:0030913) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 | 1.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 1.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.3 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 1.8 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.4 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.5 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.6 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 | 0.1 | GO:0043987 | histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 1.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 1.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:2000338 | chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.3 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.2 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 1.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0090168 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) Golgi reassembly(GO:0090168) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 | 0.3 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.7 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.2 | GO:2001214 | arterial endothelial cell differentiation(GO:0060842) positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 2.0 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.2 | GO:0060261 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 0.4 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.0 | 0.1 | GO:0061684 | chaperone-mediated autophagy(GO:0061684) |
| 0.0 | 0.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 | 0.0 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.0 | GO:2000109 | macrophage apoptotic process(GO:0071888) regulation of macrophage apoptotic process(GO:2000109) |
| 0.0 | 0.5 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 0.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.7 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.3 | 3.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.2 | 15.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.8 | 4.0 | GO:1990462 | omegasome(GO:1990462) |
| 0.7 | 2.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.6 | 2.4 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.5 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 3.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.5 | 3.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 1.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.5 | 1.4 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.4 | 4.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 1.2 | GO:0042025 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.4 | 2.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.4 | 3.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 3.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 3.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 8.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 1.1 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 2.0 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.3 | 2.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 0.8 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 2.9 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.3 | 2.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 2.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 2.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.2 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.8 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.2 | 0.9 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 1.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 3.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 0.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 2.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 0.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 1.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 2.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.2 | 1.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.9 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.1 | 1.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.4 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 11.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 6.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 2.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 4.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 2.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.6 | GO:0061700 | nuclear pore outer ring(GO:0031080) GATOR2 complex(GO:0061700) |
| 0.1 | 4.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 8.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.7 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) platelet dense granule membrane(GO:0031088) platelet dense granule(GO:0042827) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.4 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0032806 | carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 2.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.5 | 6.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 1.4 | 4.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 1.2 | 6.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.0 | 2.9 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.8 | 2.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.7 | 2.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.7 | 9.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.6 | 2.4 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.6 | 2.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.5 | 4.4 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.5 | 2.2 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.5 | 2.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.5 | 6.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 12.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.5 | 1.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 1.4 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.5 | 1.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 1.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.4 | 2.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 1.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 0.4 | 3.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.4 | 6.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 3.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.4 | 5.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.9 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.4 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 2.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.3 | 1.7 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.3 | 1.3 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 2.2 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.3 | 0.9 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.3 | 4.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 4.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 3.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.3 | 0.8 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.3 | 1.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 0.8 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 1.0 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.2 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 1.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 4.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 0.7 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.2 | 0.9 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 1.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.8 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 0.8 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.2 | 1.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.2 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.6 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.2 | 0.5 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 0.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 1.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.5 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.2 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.5 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.2 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.2 | 0.8 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 0.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.2 | 3.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 4.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 3.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.8 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 0.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.1 | 3.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 2.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 4.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 1.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.9 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 2.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 1.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.3 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.5 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 2.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 2.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 19.1 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 2.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 10.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.5 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 2.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.1 | 4.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 0.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.5 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.2 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 2.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.0 | 1.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.3 | GO:0015168 | structural constituent of eye lens(GO:0005212) glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.5 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 1.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 7.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.0 | 2.7 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 1.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 1.7 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 1.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.4 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 5.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 5.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |