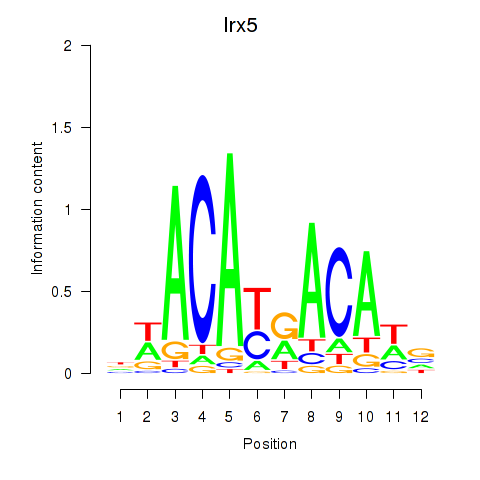
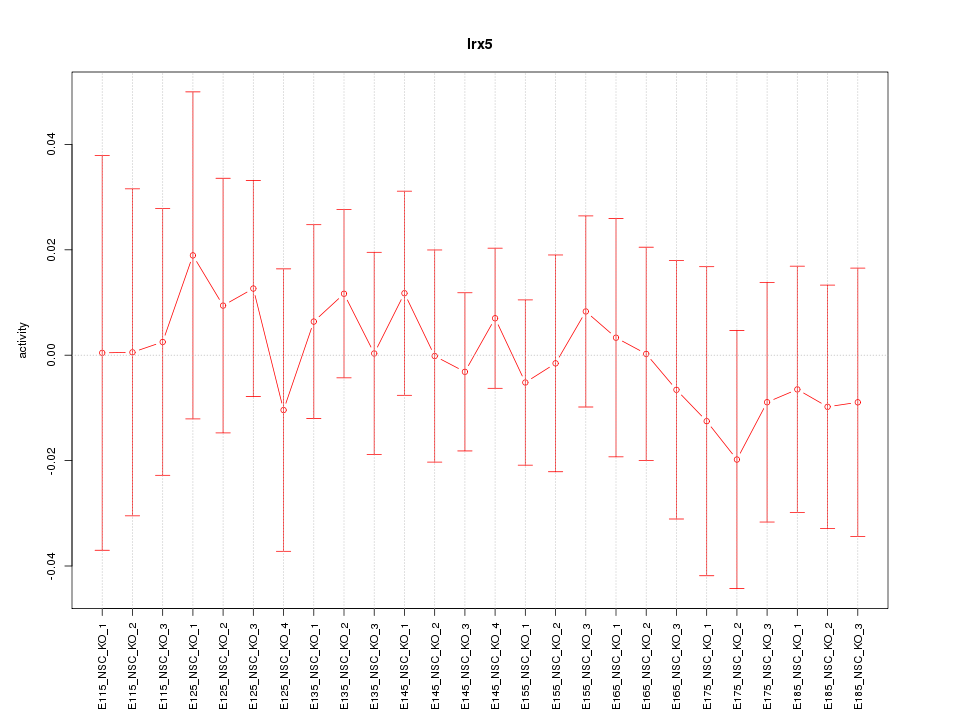
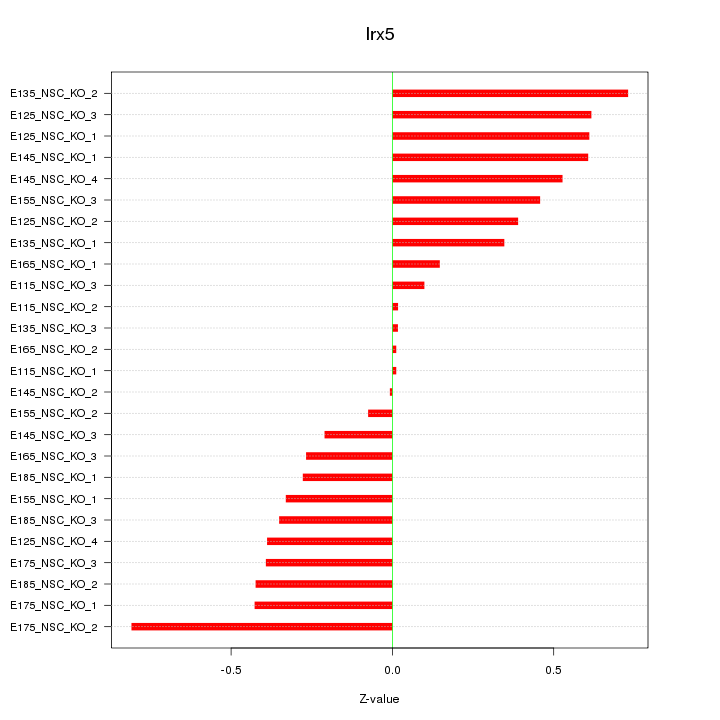
| 0.2 |
0.6 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.2 |
1.5 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
0.4 |
GO:0021557 |
oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.1 |
0.4 |
GO:0010986 |
complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 |
0.3 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.3 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 0.1 |
0.2 |
GO:1902524 |
positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.6 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.1 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.3 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 |
0.2 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.1 |
GO:2000325 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 |
0.1 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.1 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
0.3 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
0.1 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |