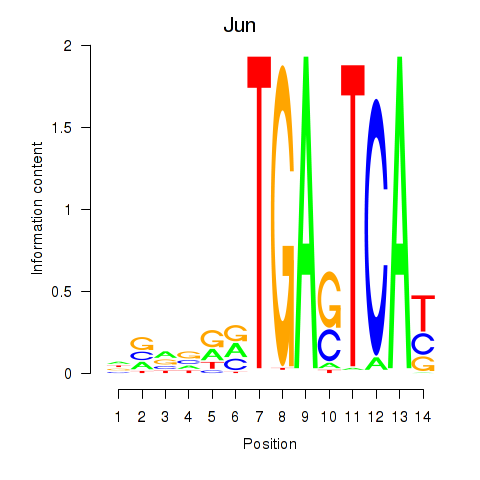
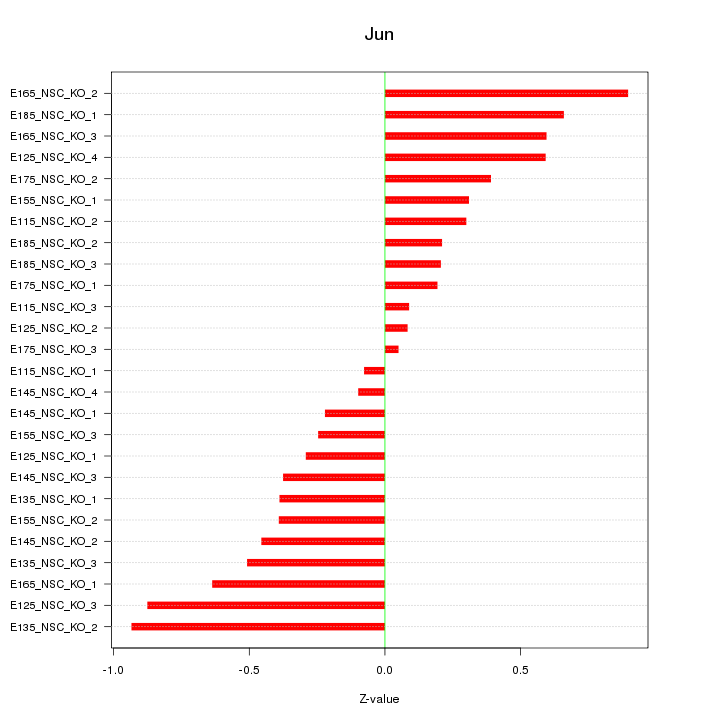
| 0.4 |
1.6 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.3 |
1.8 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.2 |
1.8 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.2 |
2.0 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.2 |
0.6 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.2 |
0.6 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.2 |
0.6 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.2 |
0.5 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.2 |
0.5 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.1 |
0.4 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 |
0.6 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.6 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.4 |
GO:0090272 |
negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.1 |
0.5 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 |
0.5 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 |
0.7 |
GO:1900029 |
positive regulation of ruffle assembly(GO:1900029) |
| 0.1 |
0.5 |
GO:0060161 |
positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 |
0.3 |
GO:0042560 |
10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 |
0.4 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.1 |
0.7 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.1 |
0.3 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.1 |
GO:0060160 |
negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 |
0.3 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
0.2 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.7 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.1 |
0.3 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 |
0.7 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.6 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.2 |
GO:0070885 |
negative regulation of interleukin-17 production(GO:0032700) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.1 |
0.3 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.4 |
GO:1903423 |
positive regulation of synaptic vesicle transport(GO:1902805) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 |
0.1 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 |
0.2 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 |
0.2 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 |
0.3 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
1.0 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.2 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.2 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.0 |
0.4 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 |
0.1 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.1 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.0 |
0.2 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.1 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.0 |
0.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.1 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 |
0.5 |
GO:0010039 |
response to iron ion(GO:0010039) |
| 0.0 |
0.3 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.0 |
0.1 |
GO:0032792 |
negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 |
0.3 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.1 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0019732 |
antifungal humoral response(GO:0019732) |
| 0.0 |
0.2 |
GO:0071594 |
T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 |
0.1 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.1 |
GO:0045084 |
positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 |
0.2 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.0 |
0.2 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.0 |
0.4 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.2 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.2 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 |
0.1 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.0 |
0.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.3 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.4 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.1 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.2 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.3 |
GO:0034113 |
heterotypic cell-cell adhesion(GO:0034113) |
| 0.0 |
0.1 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.6 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:0033198 |
response to ATP(GO:0033198) |