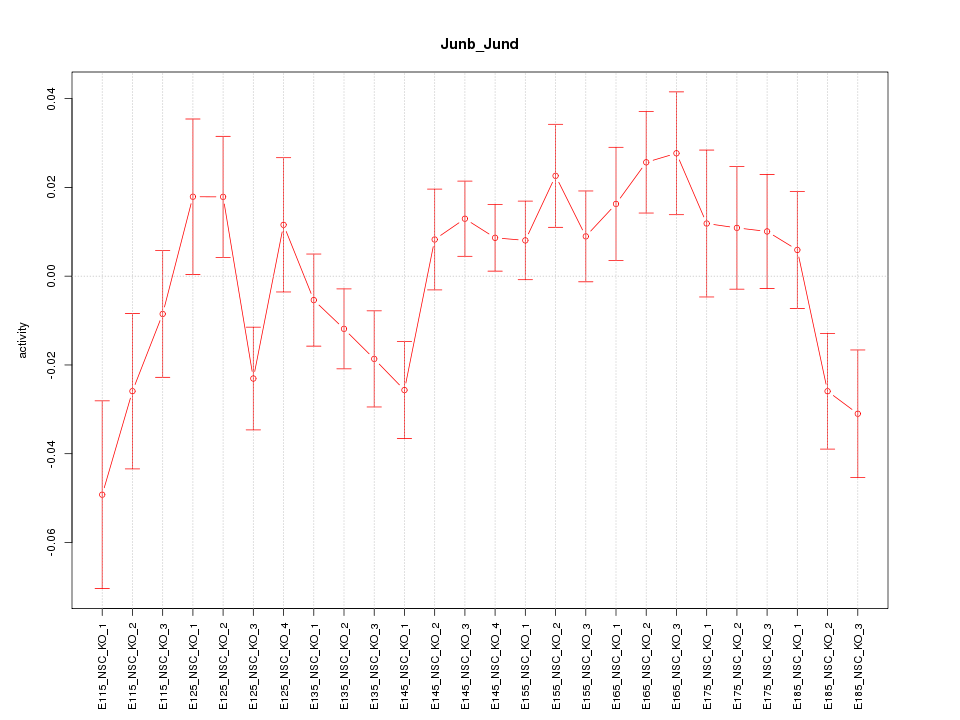
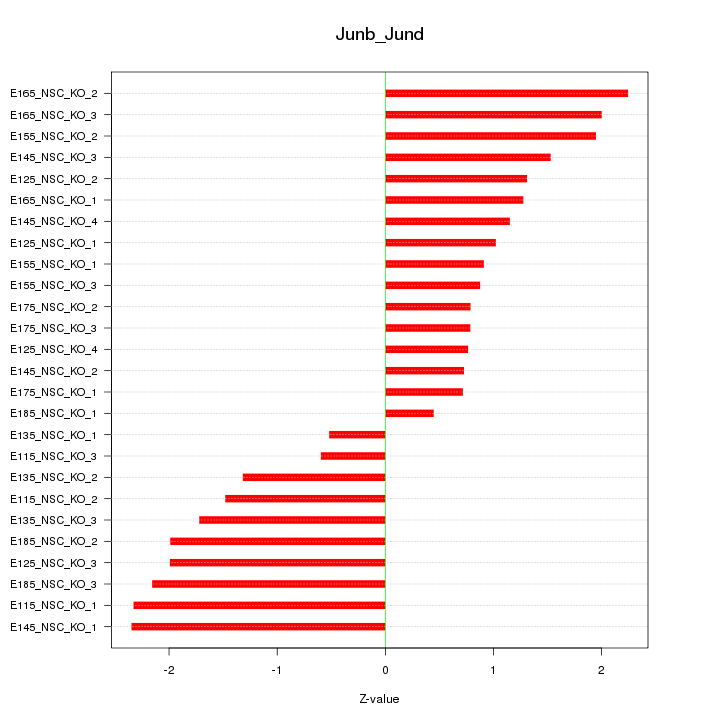
Motif ID: Junb_Jund
Z-value: 1.475
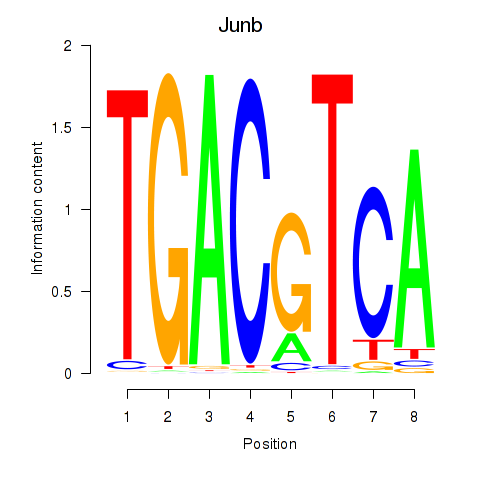
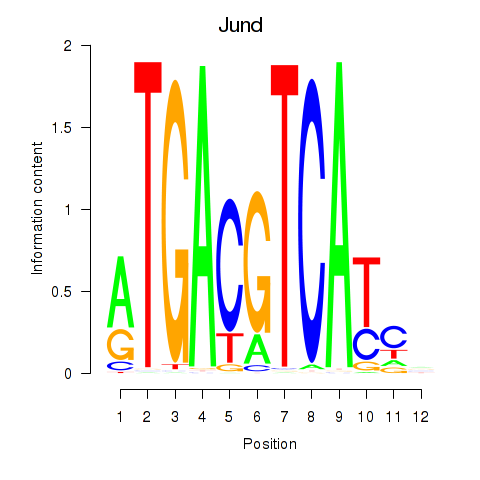
Transcription factors associated with Junb_Jund:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Junb | ENSMUSG00000052837.5 | Junb |
| Jund | ENSMUSG00000071076.5 | Jund |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Jund | mm10_v2_chr8_+_70697739_70697739 | 0.67 | 1.6e-04 | Click! |
| Junb | mm10_v2_chr8_-_84978709_84978748 | 0.58 | 1.8e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.7 | 5.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 1.3 | 5.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 1.3 | 5.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 1.2 | 5.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 1.2 | 3.5 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.1 | 3.3 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 3.2 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 1.0 | 9.2 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.0 | 4.8 | GO:0051012 | microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.9 | 2.8 | GO:0010958 | regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.9 | 2.8 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.9 | 3.6 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.9 | 3.6 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.9 | 0.9 | GO:0060067 | cervix development(GO:0060067) |
| 0.8 | 4.7 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.8 | 2.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.7 | 1.4 | GO:0035743 | CD4-positive, alpha-beta T cell cytokine production(GO:0035743) T-helper 2 cell cytokine production(GO:0035745) |
| 0.7 | 3.6 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.7 | 2.0 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.6 | 1.9 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.6 | 1.9 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.6 | 2.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.6 | 0.6 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.6 | 1.8 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.6 | 2.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.6 | 3.9 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 1.6 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.5 | 2.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 1.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.5 | 1.5 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.5 | 1.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 1.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 | 3.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 3.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.4 | 2.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 3.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.4 | 1.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.4 | 1.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.4 | 3.8 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.4 | 4.5 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.4 | 1.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.4 | 1.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.4 | 1.1 | GO:2000662 | negative regulation of interleukin-13 production(GO:0032696) interleukin-5 secretion(GO:0072603) interleukin-13 secretion(GO:0072611) regulation of interleukin-5 secretion(GO:2000662) regulation of interleukin-13 secretion(GO:2000665) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.3 | 5.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 3.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 1.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 0.9 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 | 3.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.3 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 0.9 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 1.8 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.3 | 0.9 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 2.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 0.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.3 | 2.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.3 | 4.6 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.3 | 1.7 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.3 | 2.6 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.3 | 1.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 0.8 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.3 | 1.4 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 2.8 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.3 | 0.8 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.3 | 0.8 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.3 | 1.0 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.3 | 1.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.2 | 1.0 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 1.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 3.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.2 | 0.7 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 1.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 1.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.2 | 1.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 3.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 2.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 2.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 0.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 2.0 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.2 | 1.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 0.8 | GO:1903525 | regulation of membrane tubulation(GO:1903525) |
| 0.2 | 1.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 3.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 1.0 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.2 | 1.7 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 2.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 0.9 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 11.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.2 | 7.3 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.2 | 2.0 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 3.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 13.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.2 | 1.5 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 4.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 1.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 | 3.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 0.5 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.2 | 1.5 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.4 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.1 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.6 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.1 | 2.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.6 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 2.7 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 0.7 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 1.0 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.4 | GO:0030862 | regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 | 0.8 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 1.0 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 | 0.7 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 1.0 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.8 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 1.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.5 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.1 | 0.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 3.6 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 1.4 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.3 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 1.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 4.3 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.4 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 1.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.8 | GO:0032782 | bile acid secretion(GO:0032782) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.1 | 1.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 1.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.5 | GO:1901723 | negative regulation of cell proliferation involved in kidney development(GO:1901723) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.2 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 2.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.7 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 1.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.4 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.0 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) |
| 0.1 | 0.2 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.4 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.6 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.1 | 1.1 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.1 | 0.9 | GO:0006706 | steroid catabolic process(GO:0006706) negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.3 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) response to linoleic acid(GO:0070543) |
| 0.1 | 0.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 0.2 | GO:1990314 | cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 0.1 | 0.5 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.3 | GO:1904685 | positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.1 | 0.4 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.2 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 1.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 1.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.7 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 1.6 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 2.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.7 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.1 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.1 | 0.8 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 0.5 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.1 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.7 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.1 | 1.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 2.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 3.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.7 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 3.7 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 1.8 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.5 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 3.9 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.6 | GO:0034331 | cell junction maintenance(GO:0034331) |
| 0.0 | 0.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 0.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.6 | GO:0014887 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.6 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 1.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.6 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.5 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 4.1 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.4 | GO:0044262 | cellular carbohydrate metabolic process(GO:0044262) |
| 0.0 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 1.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 2.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.7 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.7 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 1.9 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.9 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:1903299 | regulation of glucokinase activity(GO:0033131) regulation of hexokinase activity(GO:1903299) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.1 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.0 | GO:0002360 | T cell lineage commitment(GO:0002360) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 1.4 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.7 | GO:0015758 | glucose transport(GO:0015758) |
| 0.0 | 0.7 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 2.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.2 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.6 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0071451 | cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 | 0.0 | GO:1903236 | regulation of leukocyte tethering or rolling(GO:1903236) negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.0 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 20.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.7 | 3.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.7 | 10.5 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.6 | 1.9 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.6 | 3.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 3.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 4.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.5 | 2.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.5 | 5.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 2.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.4 | 1.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 1.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.4 | 1.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 7.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 1.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.3 | 1.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.3 | 9.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 1.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.3 | 12.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.6 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 4.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 0.7 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 2.9 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 1.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 1.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 3.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.9 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.2 | 5.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 2.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 8.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 2.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.1 | 2.0 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 4.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 29.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 2.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.8 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.7 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.0 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 3.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.7 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 1.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 4.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 3.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.5 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 15.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.3 | 9.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 3.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 2.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.8 | 2.3 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.7 | 2.8 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.6 | 1.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.5 | 1.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.5 | 1.6 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.5 | 1.4 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.5 | 2.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.5 | 1.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 1.9 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.5 | 2.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 1.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.5 | 1.4 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.5 | 1.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 9.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.4 | 7.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 5.1 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.3 | 1.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 11.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.3 | 1.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 3.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 1.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.3 | 3.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 0.9 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.3 | 1.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 2.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.3 | 6.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 5.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.3 | 2.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 2.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 2.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.2 | 3.7 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 1.5 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.2 | 1.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 3.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.2 | 3.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 6.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.2 | 0.4 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.2 | 2.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.2 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 2.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 2.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 0.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 0.5 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 3.1 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.2 | 1.8 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.2 | 4.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 0.5 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.2 | 5.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 0.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 2.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 3.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 11.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.7 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 5.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.6 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.9 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 3.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 3.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.1 | 6.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.7 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 4.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 3.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.1 | 2.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 3.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 4.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 4.2 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 1.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 3.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 3.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 2.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.3 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 1.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 4.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 3.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 6.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 2.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.3 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 18.3 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 5.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.5 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.0 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 3.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.2 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0045703 | pinocarveol dehydrogenase activity(GO:0018446) chloral hydrate dehydrogenase activity(GO:0018447) hydroxymethylmethylsilanediol oxidase activity(GO:0018448) 1-phenylethanol dehydrogenase activity(GO:0018449) myrtenol dehydrogenase activity(GO:0018450) cis-1,2-dihydroxy-1,2-dihydro-8-carboxynaphthalene dehydrogenase activity(GO:0034522) 3-hydroxy-4-methyloctanoyl-CoA dehydrogenase activity(GO:0034582) 2-hydroxy-4-isopropenylcyclohexane-1-carboxyl-CoA dehydrogenase activity(GO:0034778) cis-9,10-dihydroanthracene-9,10-diol dehydrogenase activity(GO:0034817) citronellol dehydrogenase activity(GO:0034821) naphthyl-2-hydroxymethyl-succinyl-CoA dehydrogenase activity(GO:0034847) 2,4,4-trimethyl-1-pentanol dehydrogenase activity(GO:0034863) 2,4,4-trimethyl-3-hydroxypentanoyl-CoA dehydrogenase activity(GO:0034868) 1-hydroxy-4,4-dimethylpentan-3-one dehydrogenase activity(GO:0034871) endosulfan diol dehydrogenase activity(GO:0034891) endosulfan hydroxyether dehydrogenase activity(GO:0034901) 3-hydroxy-2-methylhexanoyl-CoA dehydrogenase activity(GO:0034918) 3-hydroxy-2,6-dimethyl-5-methylene-heptanoyl-CoA dehydrogenase activity(GO:0034944) versicolorin reductase activity(GO:0042469) ketoreductase activity(GO:0045703) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 6.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 9.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.2 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 0.0 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 4.7 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0004497 | monooxygenase activity(GO:0004497) |