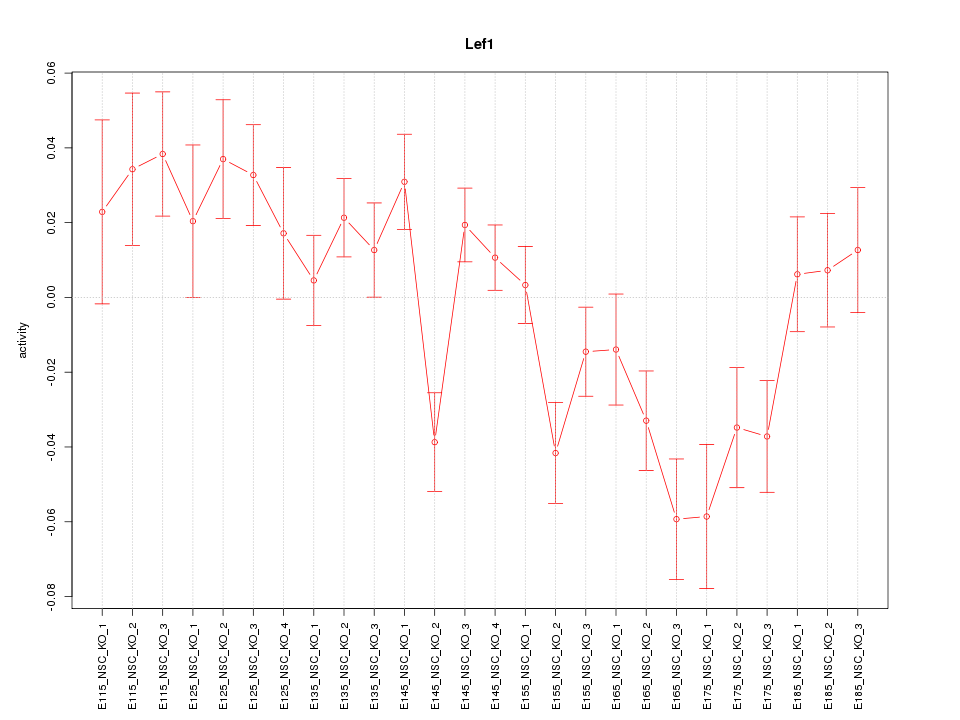
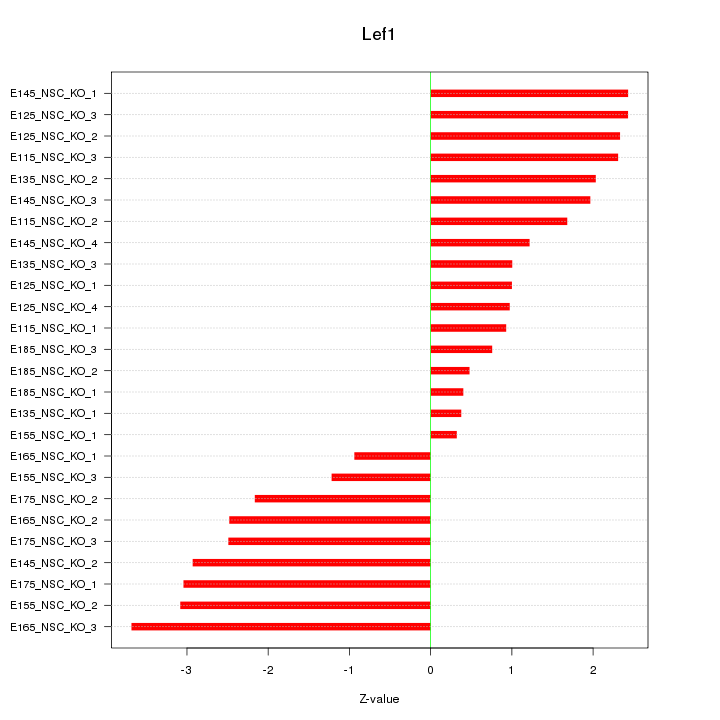
| 5.5 |
16.4 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 3.9 |
31.0 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 3.8 |
15.2 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 3.5 |
13.9 |
GO:2000054 |
negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 2.6 |
7.8 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 2.1 |
6.2 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 2.1 |
6.2 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 1.8 |
5.4 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.8 |
12.5 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.7 |
11.8 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 1.5 |
6.2 |
GO:2000705 |
regulation of dense core granule biogenesis(GO:2000705) |
| 1.4 |
4.3 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.4 |
2.8 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 1.3 |
5.1 |
GO:0007056 |
spindle assembly involved in female meiosis(GO:0007056) |
| 1.3 |
20.1 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.1 |
7.5 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 1.0 |
5.2 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 1.0 |
2.9 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.9 |
4.7 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.9 |
2.8 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.9 |
1.8 |
GO:0060242 |
contact inhibition(GO:0060242) |
| 0.8 |
2.5 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.8 |
7.3 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.8 |
4.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.8 |
3.2 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.8 |
8.3 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.7 |
3.0 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.7 |
2.2 |
GO:0090258 |
negative regulation of mitochondrial fission(GO:0090258) |
| 0.7 |
1.4 |
GO:0048619 |
embryonic hindgut morphogenesis(GO:0048619) |
| 0.7 |
3.4 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.6 |
3.9 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.6 |
7.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
1.9 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.6 |
7.6 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.6 |
3.1 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.6 |
1.9 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.6 |
3.0 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.6 |
1.8 |
GO:0060217 |
hemangioblast cell differentiation(GO:0060217) |
| 0.5 |
3.8 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.5 |
3.3 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.5 |
1.6 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 |
9.8 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.5 |
2.1 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.5 |
5.0 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.5 |
2.3 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.5 |
1.4 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.4 |
3.0 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.4 |
3.9 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.4 |
1.7 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.4 |
1.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.4 |
2.4 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 |
3.2 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.4 |
1.2 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.4 |
4.7 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 |
1.9 |
GO:2000325 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.4 |
1.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.3 |
2.8 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.3 |
6.2 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.3 |
5.5 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.3 |
1.0 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 |
7.2 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.3 |
5.2 |
GO:0045446 |
endothelial cell development(GO:0001885) endothelial cell differentiation(GO:0045446) |
| 0.3 |
6.9 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.3 |
9.0 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.3 |
1.0 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.3 |
1.3 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.3 |
1.6 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 |
2.4 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.3 |
3.0 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 |
6.2 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.3 |
1.8 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.3 |
4.5 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.2 |
1.0 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 |
0.7 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 |
2.9 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 |
1.1 |
GO:0031133 |
cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.2 |
0.7 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.2 |
2.2 |
GO:0070933 |
histone H4 deacetylation(GO:0070933) |
| 0.2 |
1.5 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.2 |
0.4 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) |
| 0.2 |
5.2 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.2 |
1.0 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
5.2 |
GO:0090132 |
epithelial cell migration(GO:0010631) tissue migration(GO:0090130) epithelium migration(GO:0090132) |
| 0.2 |
7.4 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 |
3.5 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 |
0.4 |
GO:1904468 |
negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.2 |
2.1 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.2 |
7.0 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.2 |
2.2 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 |
1.9 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.2 |
1.8 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.2 |
0.5 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.1 |
1.2 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 |
2.7 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
0.6 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
0.6 |
GO:0001865 |
NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 |
0.9 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.1 |
0.5 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
1.8 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.1 |
1.0 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.2 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
1.2 |
GO:0060712 |
spongiotrophoblast layer development(GO:0060712) |
| 0.1 |
1.2 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 |
2.0 |
GO:0060538 |
skeletal muscle organ development(GO:0060538) |
| 0.1 |
0.6 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 |
0.9 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
1.5 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.6 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
0.5 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.1 |
1.4 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.4 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 |
0.4 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
0.8 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) detoxification of inorganic compound(GO:0061687) |
| 0.1 |
2.4 |
GO:0009649 |
entrainment of circadian clock(GO:0009649) |
| 0.1 |
0.8 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 |
0.5 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
0.8 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
0.7 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.3 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 |
3.7 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 |
0.5 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) regulation of hematopoietic stem cell proliferation(GO:1902033) |
| 0.1 |
0.7 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.6 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.7 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.1 |
0.2 |
GO:0060431 |
primary lung bud formation(GO:0060431) |
| 0.1 |
1.1 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.1 |
0.8 |
GO:0043928 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 |
9.8 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.1 |
1.4 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
3.1 |
GO:0060612 |
adipose tissue development(GO:0060612) |
| 0.1 |
1.1 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
0.2 |
GO:0044337 |
canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.1 |
0.3 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) positive regulation of mesoderm development(GO:2000382) |
| 0.1 |
0.4 |
GO:0048630 |
skeletal muscle tissue growth(GO:0048630) |
| 0.1 |
1.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
1.7 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 |
2.5 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.1 |
0.1 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.1 |
2.0 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
1.4 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.1 |
0.4 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
2.6 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.1 |
0.6 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
1.6 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.1 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.0 |
0.1 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 |
2.0 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 |
0.3 |
GO:0045667 |
regulation of osteoblast differentiation(GO:0045667) |
| 0.0 |
0.9 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.4 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.0 |
0.1 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.0 |
0.9 |
GO:0042552 |
myelination(GO:0042552) |
| 0.0 |
0.6 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.6 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.4 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.6 |
GO:2000171 |
negative regulation of dendrite development(GO:2000171) |
| 0.0 |
1.9 |
GO:0030326 |
embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 |
0.2 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.5 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
1.4 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 |
1.6 |
GO:0000070 |
mitotic sister chromatid segregation(GO:0000070) |
| 0.0 |
1.0 |
GO:0032204 |
regulation of telomere maintenance(GO:0032204) |
| 0.0 |
1.4 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
1.1 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 |
0.0 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.0 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |