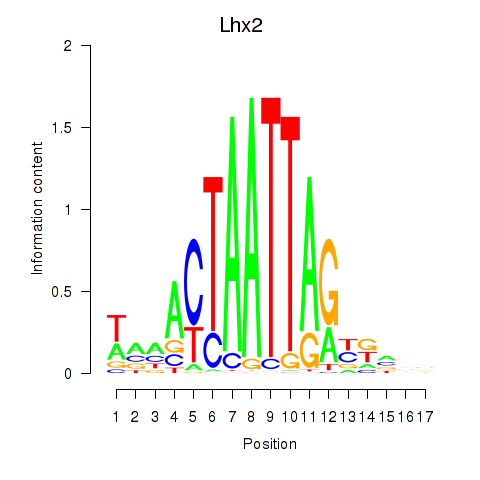
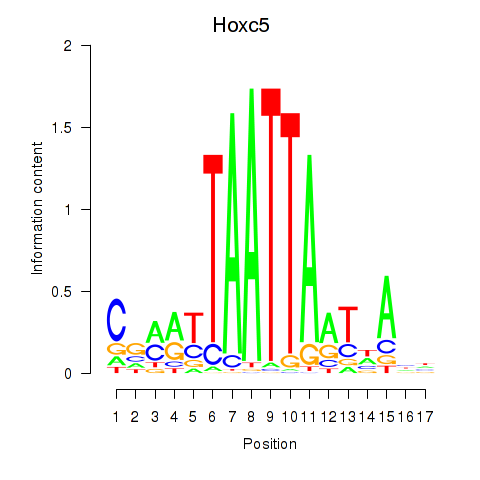
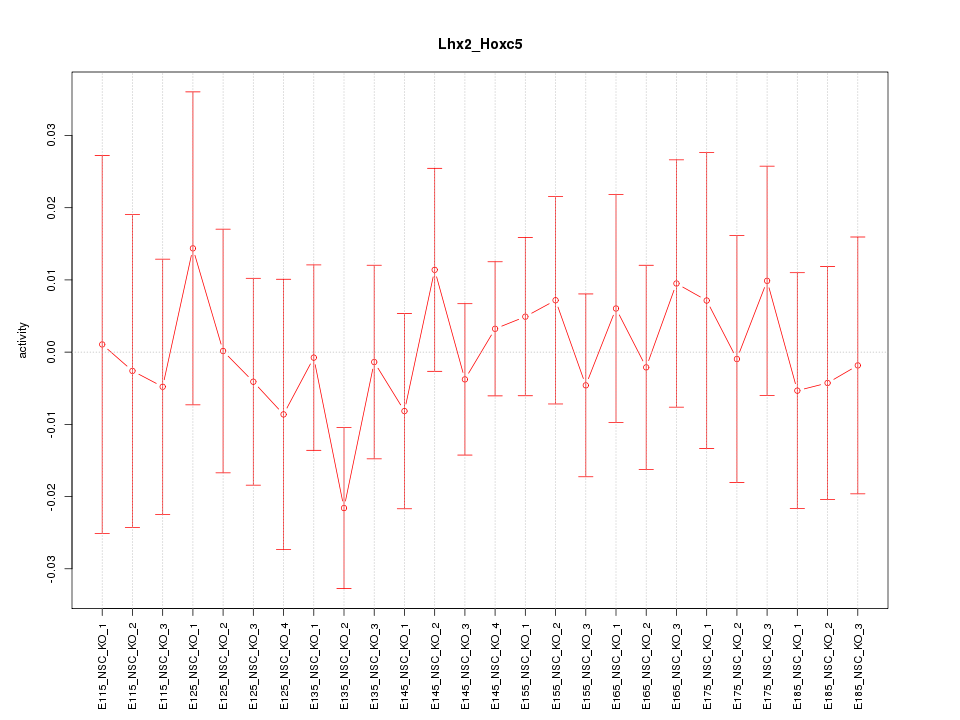
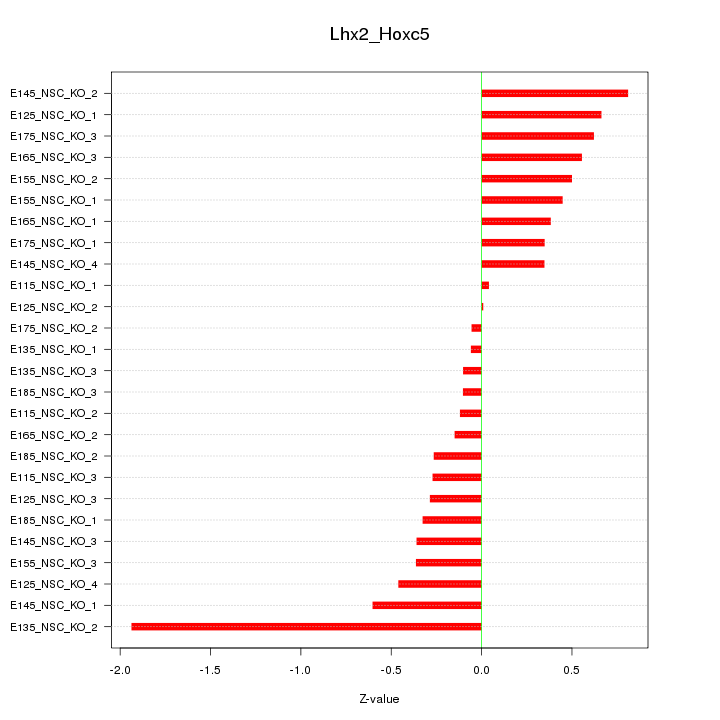
| 0.2 |
0.9 |
GO:0008627 |
intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 |
0.7 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
0.5 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
0.9 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.2 |
0.5 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.2 |
0.5 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.4 |
GO:0032700 |
negative regulation of interleukin-17 production(GO:0032700) |
| 0.1 |
1.0 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.4 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 |
0.6 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
0.4 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.5 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.1 |
0.3 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.3 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 |
0.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.3 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.3 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 |
0.3 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
0.3 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 |
0.3 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.1 |
0.4 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.1 |
0.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.4 |
GO:1902255 |
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 |
0.5 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
0.9 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.1 |
GO:0009106 |
lipoate metabolic process(GO:0009106) |
| 0.1 |
0.2 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 |
0.3 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 |
0.2 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.2 |
GO:0071726 |
response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 |
0.2 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.4 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 |
0.1 |
GO:1903351 |
response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) regulation of membrane tubulation(GO:1903525) |
| 0.0 |
0.1 |
GO:0006097 |
glyoxylate cycle(GO:0006097) |
| 0.0 |
0.9 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.2 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.4 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.0 |
0.2 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.3 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.0 |
0.1 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 |
0.3 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.2 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 |
0.2 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.4 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.2 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 |
0.0 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.1 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 |
0.2 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.4 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 |
1.4 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.0 |
0.2 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 |
0.2 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.0 |
0.3 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 |
0.2 |
GO:0042940 |
D-amino acid transport(GO:0042940) |
| 0.0 |
0.2 |
GO:0052695 |
cellular glucuronidation(GO:0052695) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
0.8 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.7 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.4 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.0 |
0.1 |
GO:0016561 |
protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 |
0.1 |
GO:0072338 |
creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 |
0.1 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.0 |
0.5 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 |
0.1 |
GO:0045627 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 |
0.1 |
GO:2001181 |
positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.3 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 |
0.0 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.0 |
0.2 |
GO:0060576 |
regulation of microvillus assembly(GO:0032534) intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.1 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 |
0.0 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 |
0.2 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.0 |
0.1 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.1 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.0 |
0.1 |
GO:1903546 |
microtubule anchoring at centrosome(GO:0034454) negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 |
0.3 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.1 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0000393 |
spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 |
0.2 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.0 |
0.0 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.0 |
0.0 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 |
0.1 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.2 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.3 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0071816 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 |
0.5 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.1 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 |
0.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.2 |
GO:2000821 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.1 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.0 |
0.6 |
GO:1903078 |
positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 |
0.6 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.0 |
0.0 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
0.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.3 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.0 |
0.1 |
GO:0070171 |
negative regulation of tooth mineralization(GO:0070171) |
| 0.0 |
0.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.0 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.0 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.0 |
0.1 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.1 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.1 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.0 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.0 |
0.1 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.0 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 |
0.1 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 |
0.1 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.1 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |