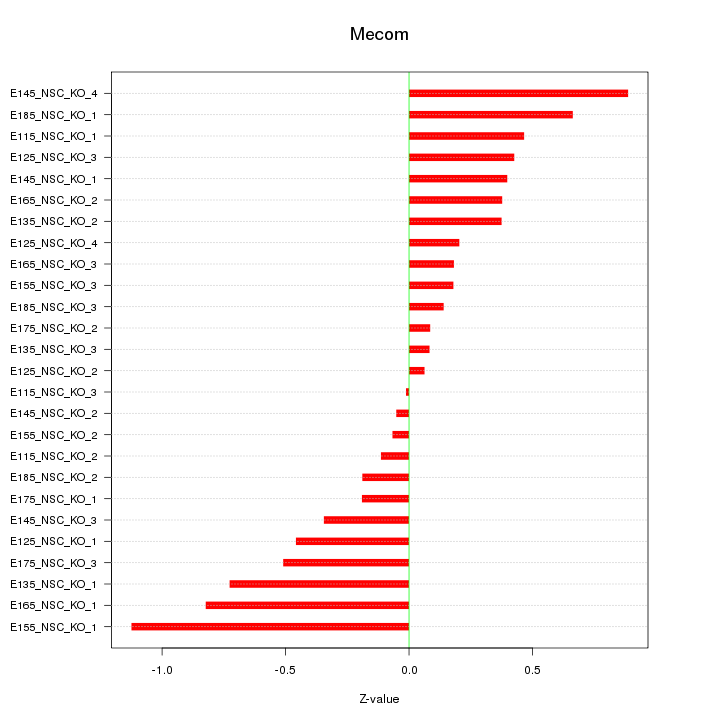
Motif ID: Mecom
Z-value: 0.454
Transcription factors associated with Mecom:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mecom | ENSMUSG00000027684.10 | Mecom |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.4 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.7 | GO:0097067 | negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 0.5 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |