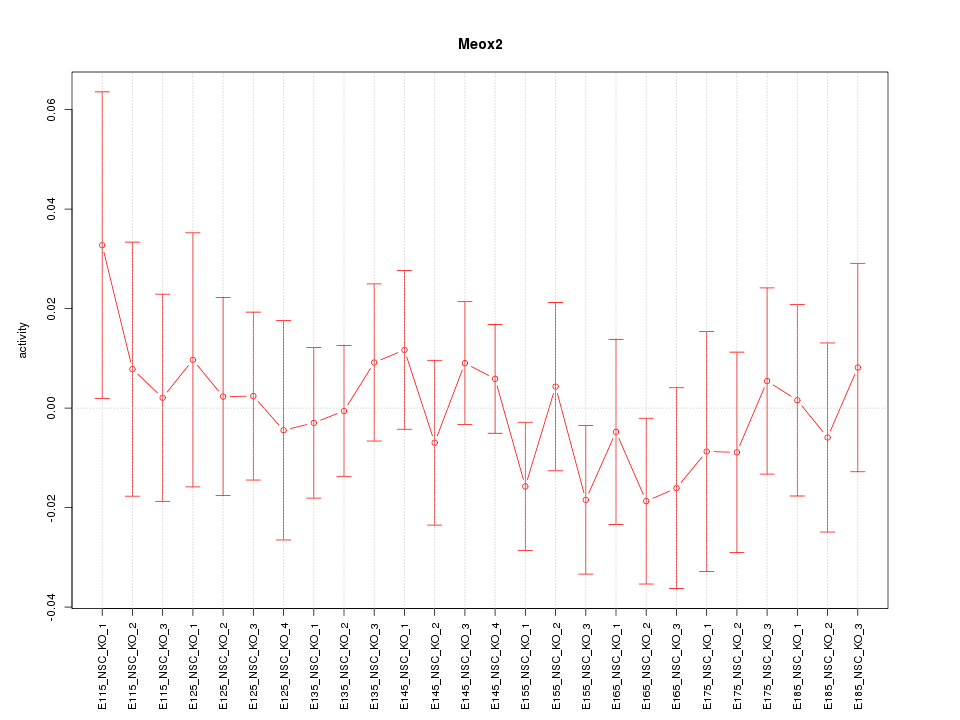
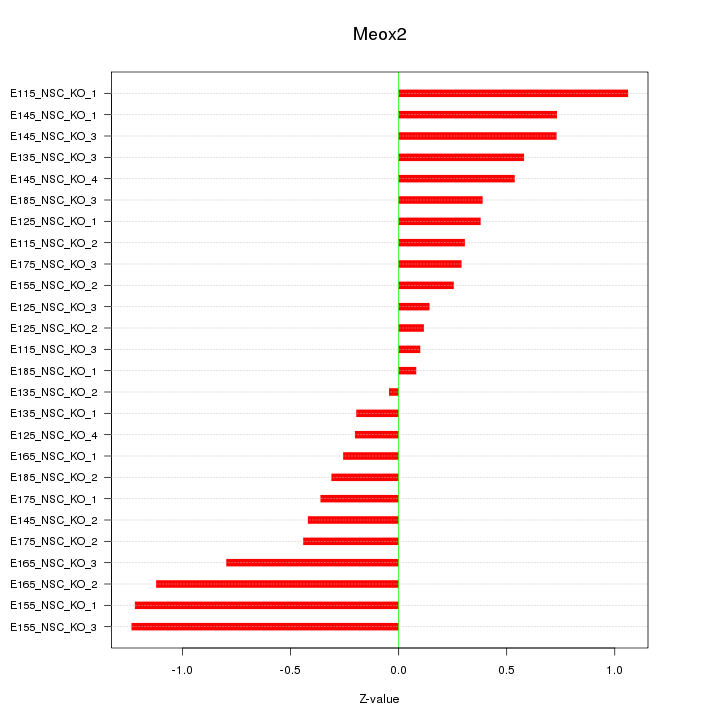
| 0.3 |
1.7 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 |
0.7 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.2 |
0.9 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 |
0.6 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.2 |
0.5 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 |
0.5 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.7 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 |
0.5 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
0.4 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
1.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.8 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.1 |
0.4 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.1 |
0.2 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 |
1.5 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.1 |
0.6 |
GO:1901970 |
positive regulation of mitotic sister chromatid separation(GO:1901970) |
| 0.1 |
0.2 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 |
0.2 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.1 |
GO:0033122 |
regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.1 |
0.4 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
0.3 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.1 |
1.9 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.1 |
0.6 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
0.7 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.3 |
GO:0009162 |
deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 |
0.5 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 |
0.8 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.6 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.4 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
0.3 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.6 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.7 |
GO:0001946 |
lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 |
0.5 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.6 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.7 |
GO:0072698 |
protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 |
0.5 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
2.7 |
GO:0045580 |
regulation of T cell differentiation(GO:0045580) |
| 0.0 |
0.2 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.0 |
0.2 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.3 |
GO:0000478 |
endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 |
0.5 |
GO:0045663 |
positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.6 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.7 |
GO:0009225 |
nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.9 |
GO:0008543 |
fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 |
0.1 |
GO:0035090 |
maintenance of apical/basal cell polarity(GO:0035090) |
| 0.0 |
0.1 |
GO:0000012 |
single strand break repair(GO:0000012) regulation of isotype switching(GO:0045191) |
| 0.0 |
0.1 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
1.0 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.1 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |