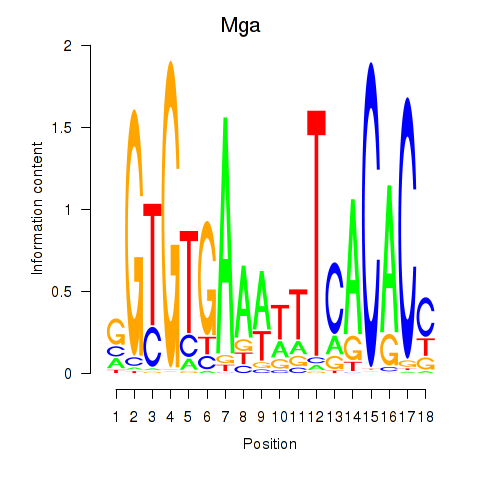
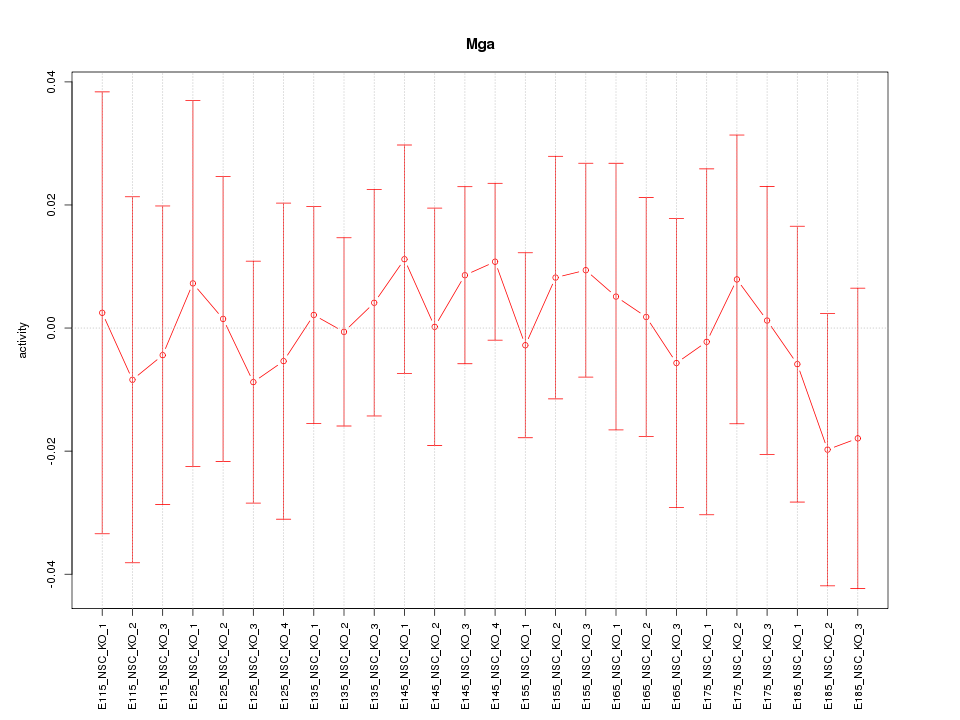
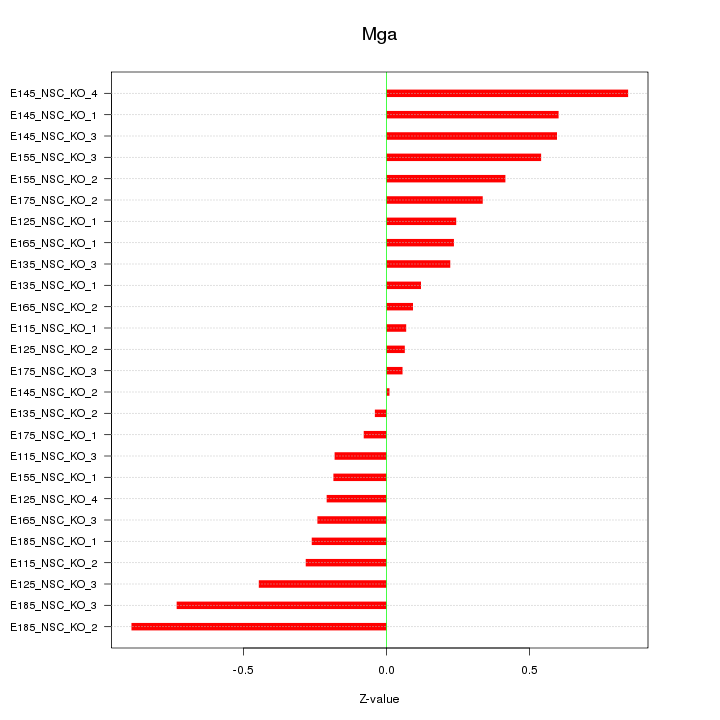
| 0.1 |
0.3 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.0 |
0.2 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.0 |
0.2 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.3 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.4 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.3 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.1 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.1 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.0 |
0.1 |
GO:0060700 |
regulation of ribonuclease activity(GO:0060700) |
| 0.0 |
0.3 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.2 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 |
0.1 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.2 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |