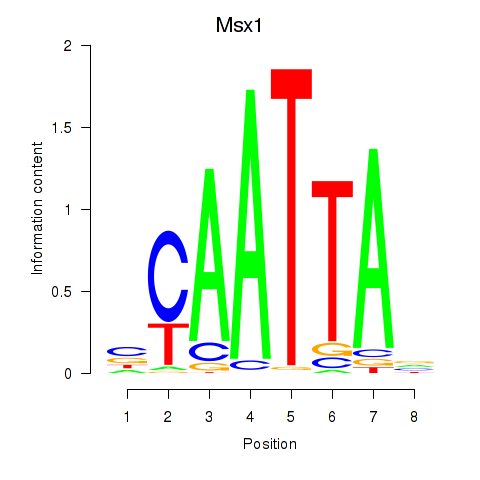
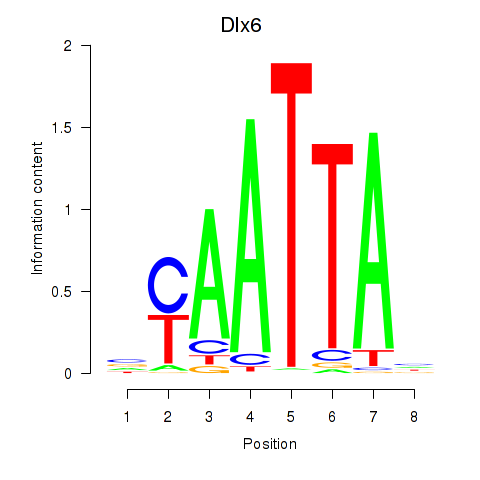
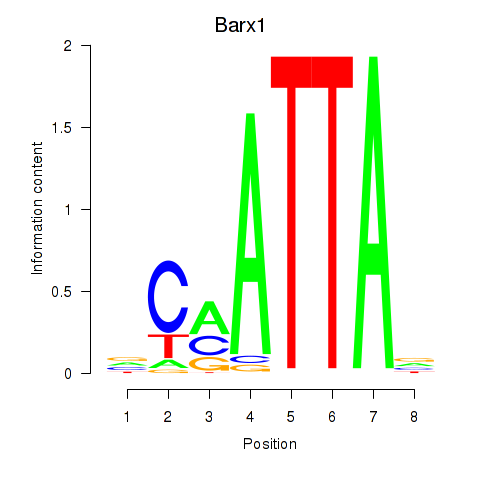
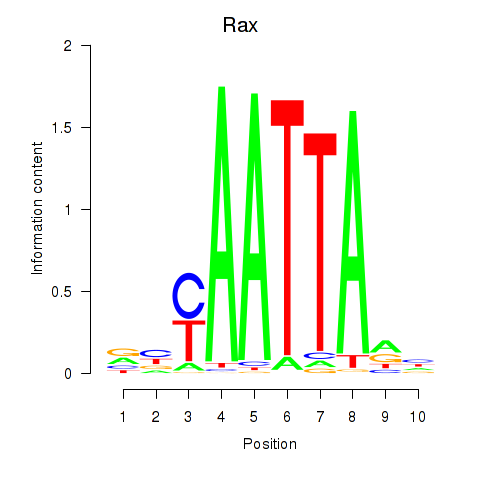
| 3.1 |
9.3 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.9 |
6.6 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.8 |
4.7 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.7 |
2.2 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.6 |
1.8 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.6 |
1.7 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.4 |
6.5 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.4 |
3.5 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.4 |
0.4 |
GO:0060915 |
fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.4 |
2.7 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.4 |
2.3 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 |
9.4 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.4 |
1.9 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 |
0.9 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.3 |
1.1 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.3 |
1.1 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.3 |
0.8 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 |
0.5 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 |
0.9 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.6 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 0.1 |
1.6 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.7 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.8 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.1 |
1.9 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.1 |
0.6 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
2.1 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.5 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
0.4 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.4 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
3.7 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.1 |
0.4 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.7 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.1 |
1.0 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.2 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) glomerular mesangial cell differentiation(GO:0072008) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.4 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 |
1.2 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
0.2 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 |
0.2 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 |
0.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.0 |
1.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
4.5 |
GO:0043149 |
contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
1.6 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.3 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
2.9 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.0 |
0.6 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
1.0 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.5 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.9 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.4 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.6 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.4 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.0 |
0.1 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.2 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.1 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.0 |
0.3 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0051482 |
positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 |
1.4 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.3 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
1.8 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.0 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |