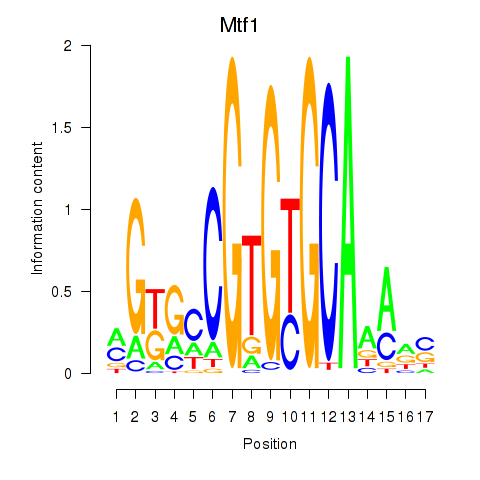
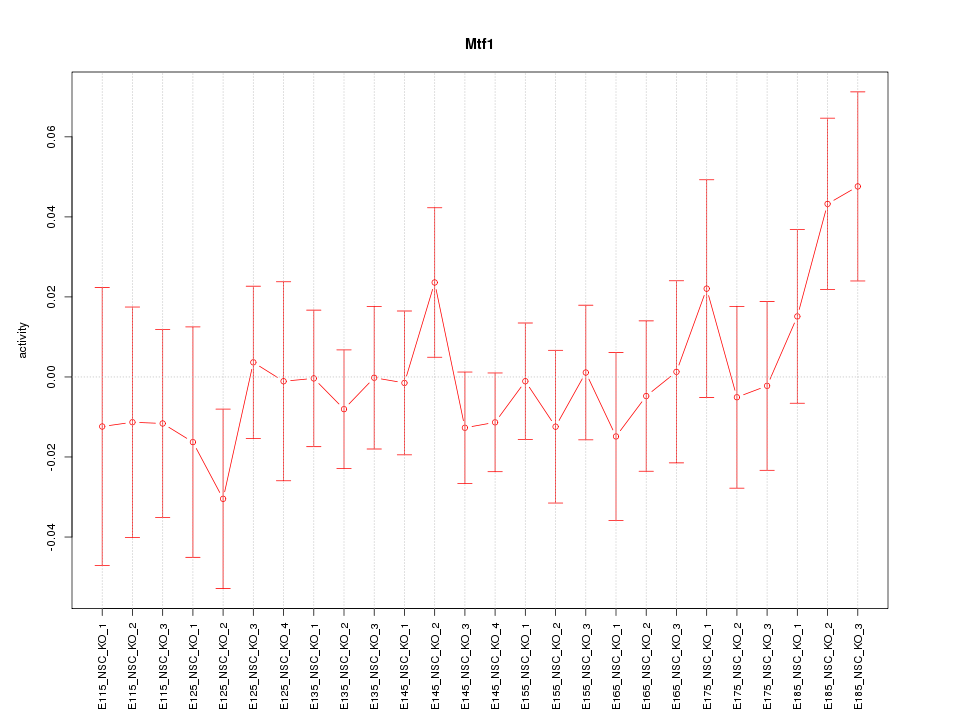
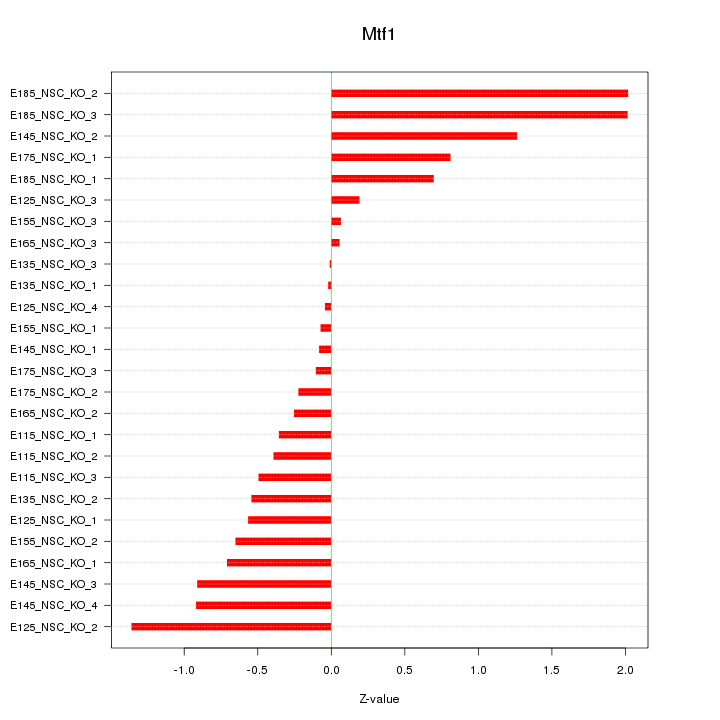
| 2.2 |
15.1 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.5 |
0.5 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) |
| 0.3 |
5.5 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
0.6 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 |
0.4 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
0.4 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.8 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 |
0.3 |
GO:0015920 |
regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.1 |
0.3 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.8 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 |
0.4 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 |
0.6 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.5 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
0.5 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.2 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 |
1.1 |
GO:0048715 |
negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.1 |
0.5 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
3.0 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
0.3 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.1 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.2 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.1 |
GO:2000275 |
regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 |
0.3 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
1.4 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.6 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 |
0.1 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.0 |
1.0 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.2 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |