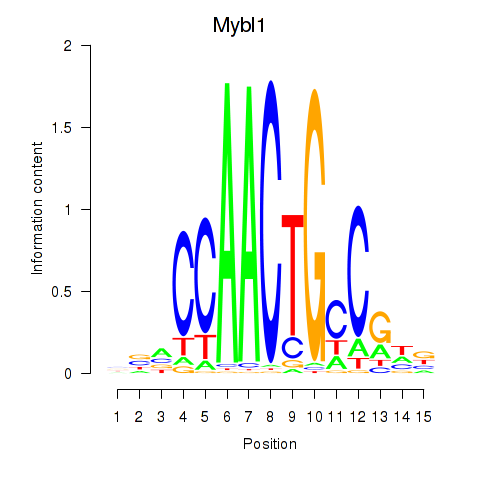
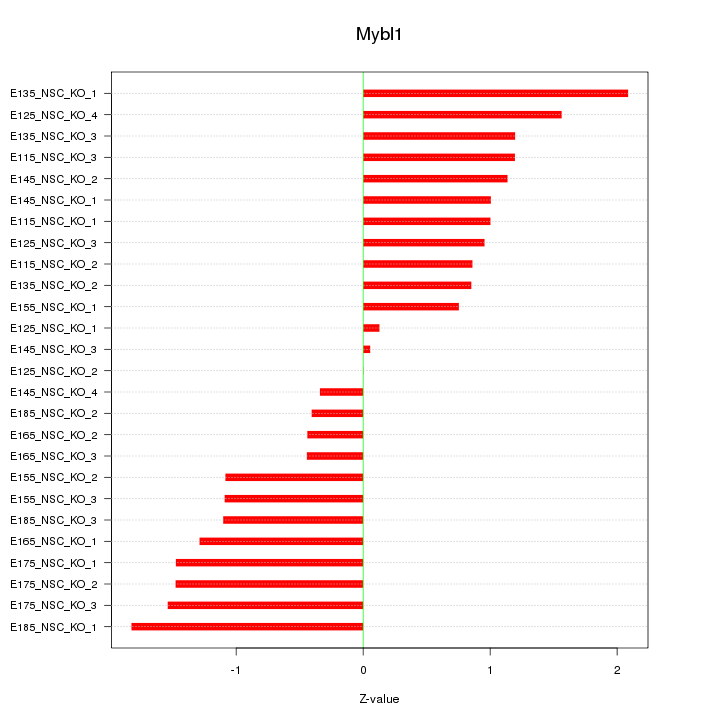
| 1.0 |
4.0 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.9 |
5.3 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.8 |
2.5 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.8 |
2.4 |
GO:0044778 |
meiotic DNA integrity checkpoint(GO:0044778) |
| 0.8 |
3.1 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.6 |
2.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.6 |
3.1 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.5 |
1.6 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.5 |
2.6 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.5 |
4.7 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.5 |
2.0 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.5 |
2.0 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.5 |
8.3 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.5 |
0.9 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.4 |
1.7 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.4 |
1.3 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.4 |
3.2 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.4 |
1.9 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.4 |
1.2 |
GO:0040009 |
regulation of growth rate(GO:0040009) protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 |
2.3 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.4 |
2.2 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 |
1.1 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.3 |
2.1 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.3 |
1.6 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 |
1.6 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 |
0.8 |
GO:0071649 |
regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.3 |
1.1 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 |
1.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.3 |
2.4 |
GO:0044458 |
motile cilium assembly(GO:0044458) |
| 0.3 |
1.3 |
GO:0090666 |
telomere assembly(GO:0032202) scaRNA localization to Cajal body(GO:0090666) |
| 0.3 |
0.8 |
GO:0006428 |
isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.3 |
1.5 |
GO:0035087 |
siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.2 |
6.1 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 |
1.9 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 |
1.7 |
GO:1902965 |
regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.2 |
2.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.2 |
0.7 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.2 |
2.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.2 |
1.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
1.1 |
GO:0010519 |
negative regulation of phospholipase activity(GO:0010519) |
| 0.2 |
0.9 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 |
0.8 |
GO:1900157 |
regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.2 |
0.6 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 |
0.6 |
GO:0019405 |
hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) fructose biosynthetic process(GO:0046370) |
| 0.2 |
0.9 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.2 |
0.7 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.2 |
0.5 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.2 |
0.5 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 |
1.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.2 |
0.5 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.2 |
0.5 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.7 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 |
2.4 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.2 |
0.5 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 |
1.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.2 |
2.4 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 |
1.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 |
0.5 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.2 |
0.6 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.4 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 |
0.7 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.1 |
0.4 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 |
2.5 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.1 |
0.4 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 |
0.8 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 |
0.5 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.3 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
1.1 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.1 |
0.7 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.7 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
0.4 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 |
0.3 |
GO:0032714 |
negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 |
0.7 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.3 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 |
0.6 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
1.6 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.1 |
0.4 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.5 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 |
1.8 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
6.3 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 |
3.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.5 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 |
1.3 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.1 |
0.3 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.1 |
0.6 |
GO:0031860 |
telomeric 3' overhang formation(GO:0031860) |
| 0.1 |
0.3 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
0.6 |
GO:0045667 |
regulation of osteoblast differentiation(GO:0045667) |
| 0.1 |
0.2 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 |
0.6 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.1 |
0.8 |
GO:0000491 |
small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.1 |
0.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 |
1.1 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.1 |
0.5 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 |
1.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.2 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) |
| 0.1 |
1.9 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 |
0.3 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
0.4 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 |
0.6 |
GO:0046015 |
regulation of transcription by glucose(GO:0046015) |
| 0.1 |
0.7 |
GO:0006363 |
termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
0.7 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 |
0.6 |
GO:1902036 |
regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 |
0.8 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 |
0.3 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 |
1.4 |
GO:0046599 |
regulation of centriole replication(GO:0046599) |
| 0.1 |
0.3 |
GO:0061188 |
regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 |
2.5 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
0.4 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.4 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
0.8 |
GO:0046060 |
dATP metabolic process(GO:0046060) |
| 0.1 |
0.2 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.6 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.7 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 |
0.4 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
3.1 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
1.0 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.1 |
1.8 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 |
0.7 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.1 |
1.0 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.2 |
GO:1901675 |
response to methylglyoxal(GO:0051595) negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 |
1.0 |
GO:0001711 |
endodermal cell fate commitment(GO:0001711) |
| 0.1 |
0.4 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 |
0.4 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.7 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
1.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.1 |
1.6 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
0.7 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 |
0.7 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.1 |
0.1 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) regulation of vitamin D receptor signaling pathway(GO:0070562) cellular response to vitamin D(GO:0071305) |
| 0.1 |
0.8 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.1 |
0.4 |
GO:0010454 |
negative regulation of cell fate commitment(GO:0010454) |
| 0.1 |
0.3 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.5 |
GO:0002155 |
regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 |
0.3 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.4 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.1 |
0.8 |
GO:0007080 |
mitotic metaphase plate congression(GO:0007080) |
| 0.1 |
0.5 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 |
2.7 |
GO:0032435 |
negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.1 |
0.7 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
1.3 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
1.3 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.1 |
0.2 |
GO:0051771 |
negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 |
0.2 |
GO:0035166 |
post-embryonic hemopoiesis(GO:0035166) |
| 0.0 |
0.3 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
0.1 |
GO:0046671 |
negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 |
2.8 |
GO:0051384 |
response to glucocorticoid(GO:0051384) |
| 0.0 |
0.4 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
1.0 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.4 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
0.1 |
GO:0021592 |
fourth ventricle development(GO:0021592) |
| 0.0 |
0.1 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 |
0.8 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
1.4 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.5 |
GO:0032515 |
negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 |
1.4 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.4 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.5 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) |
| 0.0 |
0.7 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.0 |
0.6 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.9 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.1 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.0 |
0.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.2 |
GO:0009169 |
purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 |
0.3 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 |
0.3 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.4 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.6 |
GO:0006516 |
glycoprotein catabolic process(GO:0006516) |
| 0.0 |
0.8 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.0 |
0.4 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.8 |
GO:1902042 |
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 |
0.5 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.4 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.0 |
1.3 |
GO:0042273 |
ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 |
1.1 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.0 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 |
9.4 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
0.1 |
GO:0052428 |
modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
1.1 |
GO:0042632 |
cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 |
0.8 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.3 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.0 |
0.2 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.5 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.7 |
GO:0010883 |
regulation of lipid storage(GO:0010883) |
| 0.0 |
0.4 |
GO:0016226 |
iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 |
0.2 |
GO:0007099 |
centriole replication(GO:0007099) |
| 0.0 |
0.5 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0060746 |
parental behavior(GO:0060746) |
| 0.0 |
0.1 |
GO:0035358 |
regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035358) positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 |
0.3 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.4 |
GO:0046621 |
negative regulation of organ growth(GO:0046621) |
| 0.0 |
0.2 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.0 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) gamma-delta T cell activation(GO:0046629) |
| 0.0 |
0.2 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.0 |
0.1 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.1 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.0 |
0.8 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
1.2 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
0.8 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.0 |
0.2 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.3 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.2 |
GO:0016926 |
protein desumoylation(GO:0016926) |
| 0.0 |
0.6 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.2 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.4 |
GO:0097194 |
execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.1 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 |
0.7 |
GO:0008625 |
extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 |
0.6 |
GO:0043303 |
mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 |
0.7 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.1 |
GO:2000059 |
regulation of ephrin receptor signaling pathway(GO:1901187) negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.5 |
GO:0003016 |
respiratory system process(GO:0003016) |
| 0.0 |
1.4 |
GO:0007286 |
spermatid development(GO:0007286) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.3 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.0 |
0.7 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) cellular oxidant detoxification(GO:0098869) |
| 0.0 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
1.5 |
GO:0008033 |
tRNA processing(GO:0008033) |
| 0.0 |
0.4 |
GO:0016445 |
somatic diversification of immunoglobulins(GO:0016445) |
| 0.0 |
0.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.1 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.1 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
0.3 |
GO:0006094 |
gluconeogenesis(GO:0006094) |
| 0.0 |
0.2 |
GO:0044070 |
regulation of anion transport(GO:0044070) |
| 0.0 |
0.2 |
GO:0017144 |
drug metabolic process(GO:0017144) |
| 0.0 |
0.0 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.5 |
GO:0071594 |
T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 |
0.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |