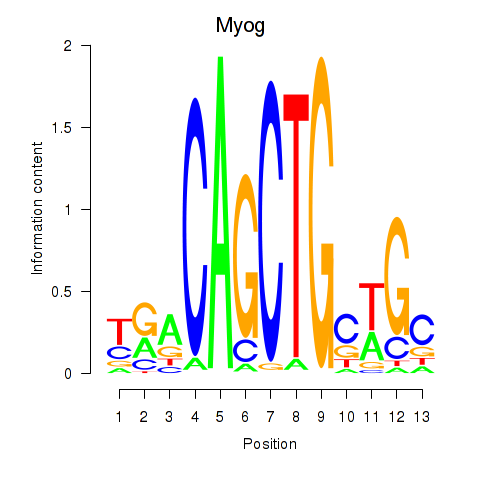
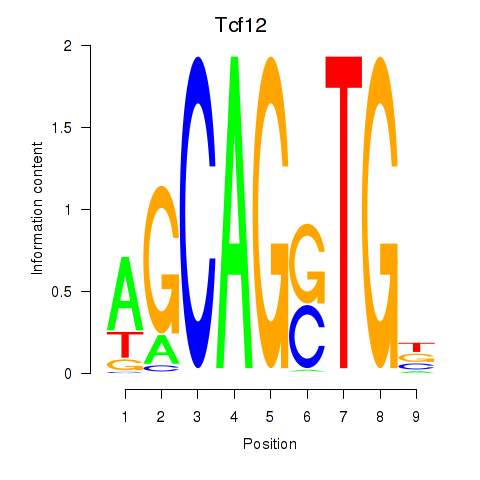
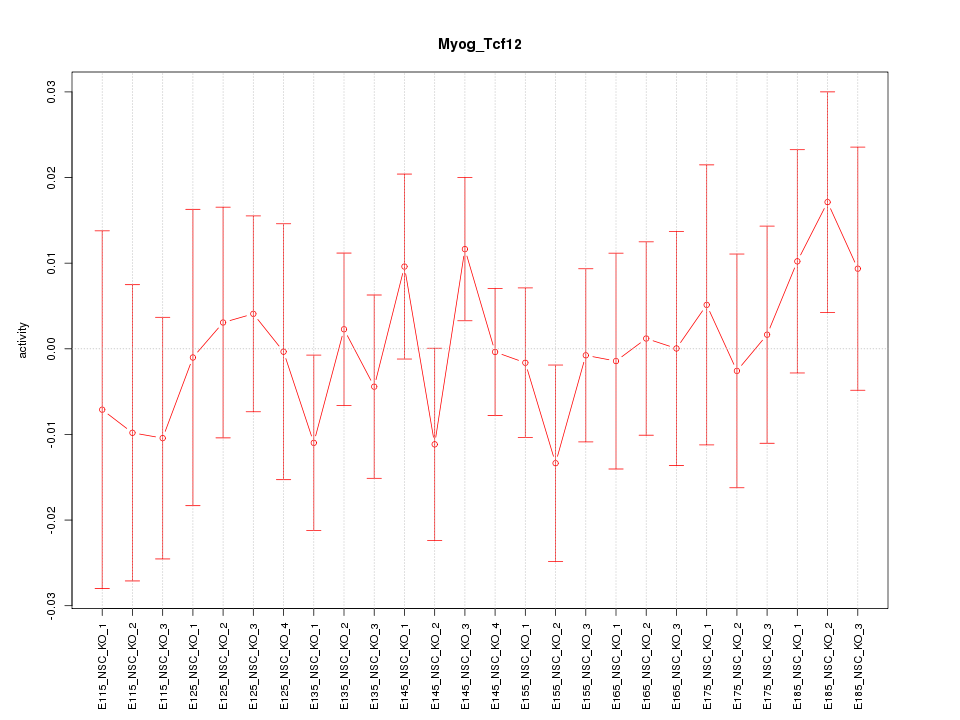
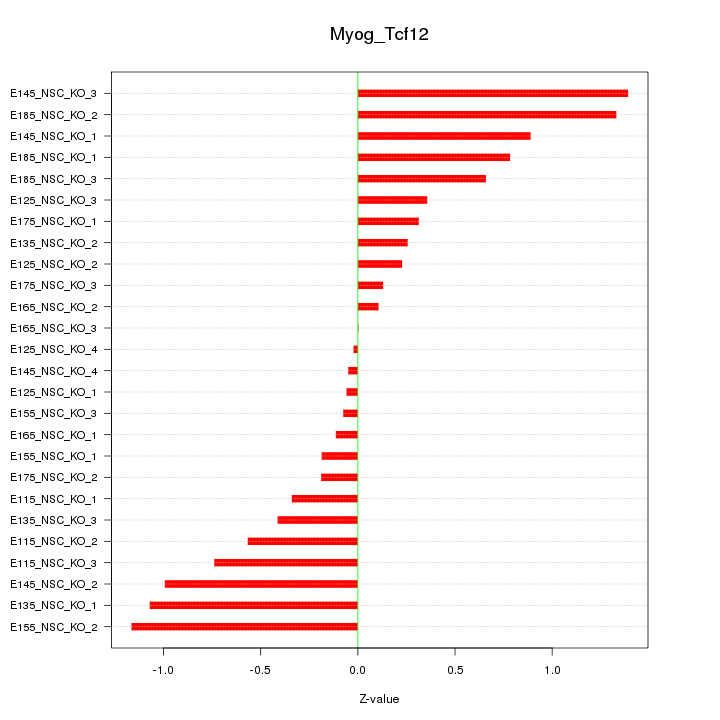
| 0.9 |
3.4 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 |
3.4 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 |
0.4 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 |
1.1 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 0.3 |
1.4 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.3 |
1.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 |
0.9 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.3 |
1.8 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 |
0.9 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.3 |
0.9 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 |
0.9 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.3 |
1.1 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.3 |
0.8 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 |
1.6 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 |
1.3 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 |
0.8 |
GO:0007521 |
muscle cell fate determination(GO:0007521) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 |
1.6 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 |
0.6 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 |
0.6 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.2 |
0.6 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.2 |
0.6 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.2 |
0.4 |
GO:1903275 |
positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.2 |
1.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 |
0.5 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.2 |
0.3 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 |
0.6 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.4 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
1.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
0.4 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.4 |
GO:0046959 |
habituation(GO:0046959) |
| 0.1 |
0.6 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 |
0.5 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
0.2 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.5 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.3 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 |
0.5 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.5 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.1 |
0.5 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.4 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.2 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
0.6 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.2 |
GO:0002513 |
tolerance induction to self antigen(GO:0002513) |
| 0.1 |
0.5 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.1 |
0.8 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
0.4 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
0.6 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
1.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.1 |
0.2 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.3 |
GO:1900740 |
positive regulation of thymocyte apoptotic process(GO:0070245) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.1 |
GO:0032714 |
negative regulation of interleukin-13 production(GO:0032696) negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 |
0.2 |
GO:1905154 |
negative regulation of membrane invagination(GO:1905154) |
| 0.1 |
0.5 |
GO:1904017 |
response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) regulation of response to drug(GO:2001023) |
| 0.1 |
0.5 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.2 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.4 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 |
0.2 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.4 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.1 |
0.6 |
GO:2000809 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.4 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
0.2 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.1 |
0.2 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.1 |
0.8 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.1 |
0.3 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
0.4 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.1 |
0.3 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 |
0.5 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.3 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
0.4 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.1 |
0.3 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) eye pigmentation(GO:0048069) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
2.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.1 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.1 |
0.2 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
0.2 |
GO:2000851 |
positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 |
0.5 |
GO:1901629 |
regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 |
0.2 |
GO:0036023 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 |
0.3 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 |
0.2 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
0.2 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
2.3 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.1 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
2.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.2 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.1 |
0.2 |
GO:0060452 |
positive regulation of cardiac muscle contraction(GO:0060452) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 |
1.0 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.2 |
GO:0060912 |
cardiac cell fate specification(GO:0060912) |
| 0.1 |
0.3 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.1 |
0.3 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.5 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.2 |
GO:0042636 |
striatal medium spiny neuron differentiation(GO:0021773) negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.0 |
0.2 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
1.3 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.7 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.0 |
0.2 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.0 |
0.1 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 |
0.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.0 |
0.8 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 |
0.4 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
1.2 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.0 |
0.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 |
0.4 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.2 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 |
0.2 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 |
0.2 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.6 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.0 |
0.1 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.0 |
0.1 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.0 |
0.7 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.2 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.0 |
0.1 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.2 |
GO:0060923 |
muscle cell fate commitment(GO:0042693) cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 |
0.1 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.0 |
0.6 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.3 |
GO:0006930 |
substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 |
0.1 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 |
0.6 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.3 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 |
0.1 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.2 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.0 |
0.2 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.2 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.2 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 |
0.2 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 |
0.1 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.0 |
0.1 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 |
0.1 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.0 |
0.1 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 |
0.1 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.5 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
1.0 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.0 |
0.1 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.3 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.7 |
GO:1901522 |
positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 |
0.1 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 |
0.1 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) late viral transcription(GO:0019086) |
| 0.0 |
0.1 |
GO:0061535 |
glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 |
0.1 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.0 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.0 |
0.5 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.3 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.1 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 |
0.1 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.2 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 |
0.2 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.8 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.0 |
GO:0061309 |
cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) |
| 0.0 |
0.3 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.0 |
0.3 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
0.0 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.1 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.0 |
0.2 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.1 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.0 |
0.4 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.4 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.0 |
0.1 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.3 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.0 |
0.2 |
GO:0007191 |
adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 |
0.1 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 |
0.4 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.2 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.0 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.0 |
0.1 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 |
1.1 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.3 |
GO:2000794 |
regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 |
0.3 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.0 |
0.4 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 |
0.1 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.0 |
0.2 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 |
0.0 |
GO:0021861 |
forebrain radial glial cell differentiation(GO:0021861) |
| 0.0 |
0.4 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 |
0.1 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.0 |
1.7 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.0 |
0.1 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.2 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.2 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
0.2 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.2 |
GO:0051642 |
centrosome localization(GO:0051642) |
| 0.0 |
0.2 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.0 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
0.1 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.2 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.3 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:1902047 |
polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 |
0.1 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.1 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 |
0.8 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.0 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 |
0.6 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
0.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.2 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.2 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.0 |
0.2 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.8 |
GO:0014013 |
regulation of gliogenesis(GO:0014013) |
| 0.0 |
0.1 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 |
0.0 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.0 |
0.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.2 |
GO:1902624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) positive regulation of neutrophil migration(GO:1902624) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.0 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.0 |
0.2 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.1 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.1 |
GO:1900048 |
positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 |
0.1 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.1 |
GO:0071476 |
hypotonic response(GO:0006971) cellular hypotonic response(GO:0071476) |
| 0.0 |
0.0 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 |
0.1 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.0 |
GO:1900060 |
negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 |
0.1 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.1 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.0 |
0.1 |
GO:0003157 |
endocardium development(GO:0003157) |
| 0.0 |
0.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.0 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 |
0.2 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.3 |
GO:0060122 |
inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 |
1.0 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
1.4 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
0.1 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
0.5 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.0 |
0.1 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.1 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.1 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.1 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.0 |
0.8 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.2 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.1 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.2 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.0 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.0 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 |
0.1 |
GO:0014002 |
astrocyte development(GO:0014002) |