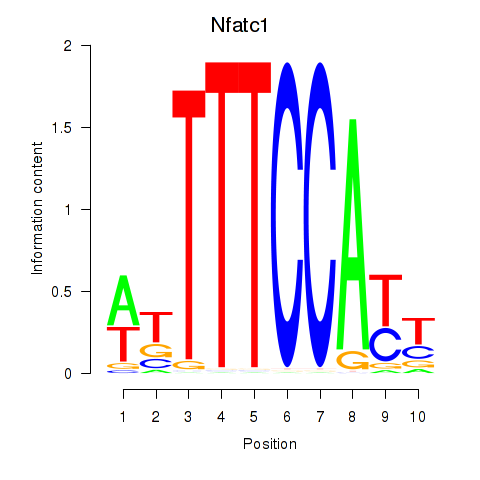
| 1.4 |
4.3 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.8 |
2.3 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.7 |
2.1 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.6 |
1.9 |
GO:0007521 |
muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.5 |
6.1 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 |
1.9 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.5 |
1.8 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.4 |
1.6 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.4 |
3.6 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.4 |
1.8 |
GO:0001575 |
globoside metabolic process(GO:0001575) |
| 0.3 |
1.0 |
GO:0043379 |
memory T cell differentiation(GO:0043379) |
| 0.3 |
1.1 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 |
1.6 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 |
2.1 |
GO:0046645 |
positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.2 |
1.7 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
0.6 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.2 |
1.4 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.2 |
0.8 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 |
0.6 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.2 |
0.5 |
GO:1902044 |
regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.2 |
1.1 |
GO:0051461 |
protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
1.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.1 |
0.5 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.6 |
GO:0007320 |
insemination(GO:0007320) |
| 0.1 |
0.7 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.1 |
0.8 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.1 |
0.5 |
GO:0072488 |
ammonium transmembrane transport(GO:0072488) |
| 0.1 |
0.6 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.2 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 |
1.0 |
GO:0045954 |
positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.1 |
0.4 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 |
0.3 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 |
0.4 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.1 |
0.4 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.7 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.1 |
0.3 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) trigeminal ganglion development(GO:0061551) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
0.7 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.7 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.1 |
1.1 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.1 |
0.4 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.1 |
0.3 |
GO:0070427 |
nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 |
0.6 |
GO:0006271 |
DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 |
0.2 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 |
0.2 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.0 |
1.5 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
4.3 |
GO:0006497 |
protein lipidation(GO:0006497) |
| 0.0 |
0.4 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.5 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.7 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.2 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.4 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.3 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.3 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.7 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.3 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.0 |
0.4 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.8 |
GO:0072348 |
sulfur compound transport(GO:0072348) |
| 0.0 |
0.5 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.0 |
0.5 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.0 |
1.0 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.4 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.2 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.4 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 |
0.1 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.5 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.6 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.3 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.8 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
341.5 |
GO:0008150 |
biological_process(GO:0008150) |