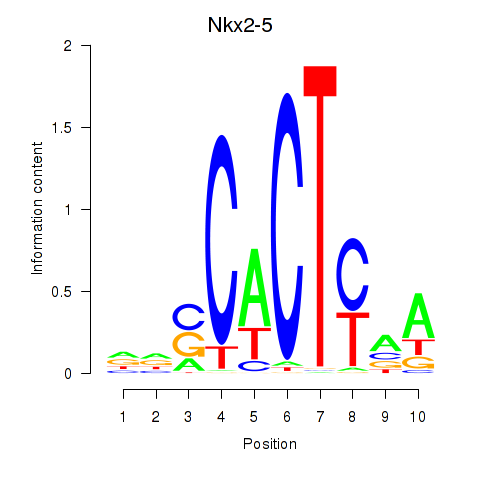
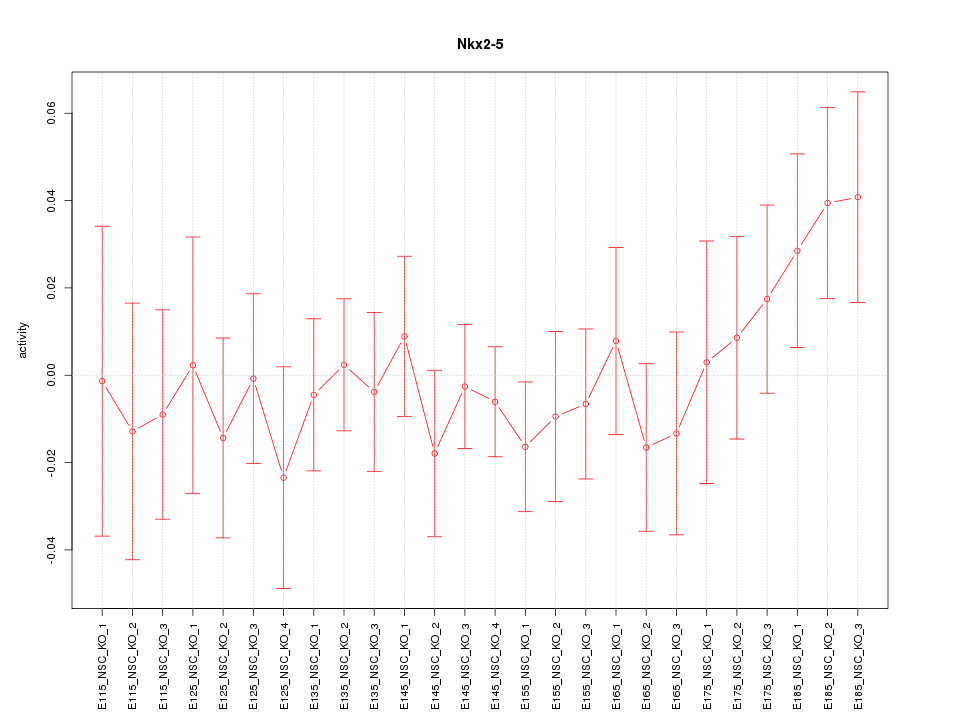
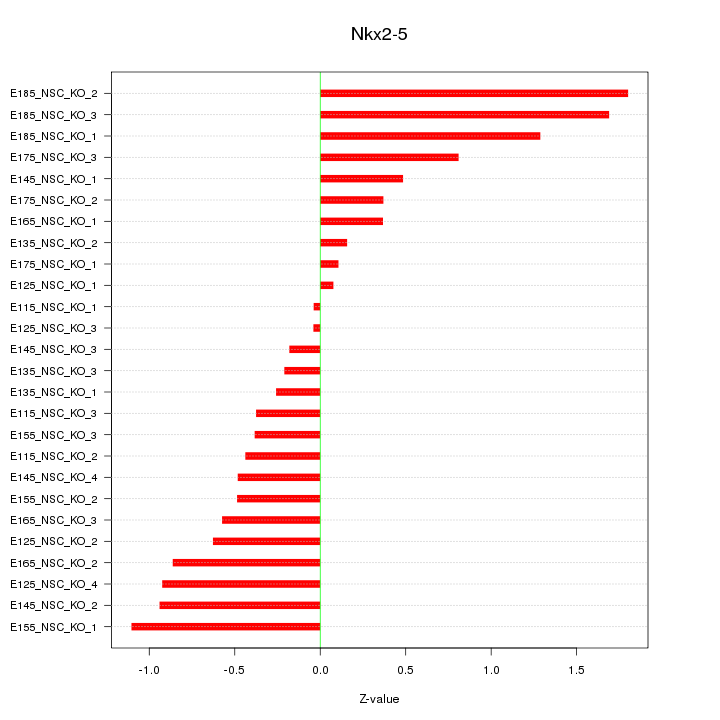
| 0.7 |
3.5 |
GO:0047484 |
regulation of response to osmotic stress(GO:0047484) |
| 0.6 |
1.9 |
GO:0051933 |
amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.4 |
3.6 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.3 |
2.4 |
GO:0034638 |
phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 |
0.7 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.2 |
1.5 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 |
0.6 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
1.6 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
2.0 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.1 |
0.4 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.9 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.8 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.4 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.1 |
1.1 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.4 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.3 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.2 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.1 |
0.5 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 |
0.7 |
GO:0051791 |
medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 |
0.3 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.2 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
1.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.0 |
0.6 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.1 |
GO:0090069 |
regulation of ribosome biogenesis(GO:0090069) |
| 0.0 |
0.3 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 |
1.3 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.3 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.6 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0060830 |
ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 |
0.4 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.1 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 |
0.4 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.8 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.4 |
GO:0006099 |
tricarboxylic acid cycle(GO:0006099) |
| 0.0 |
0.2 |
GO:2000381 |
negative regulation of mesoderm development(GO:2000381) |
| 0.0 |
0.4 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.1 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |