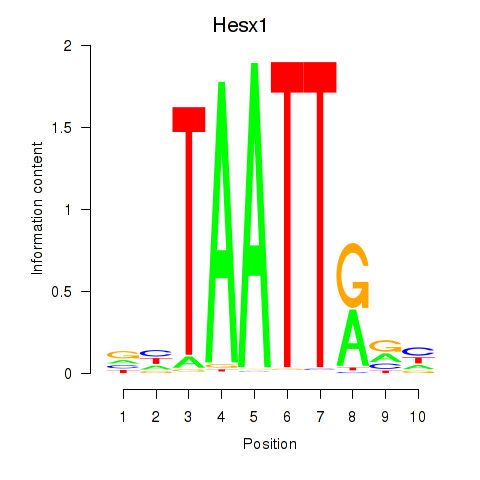
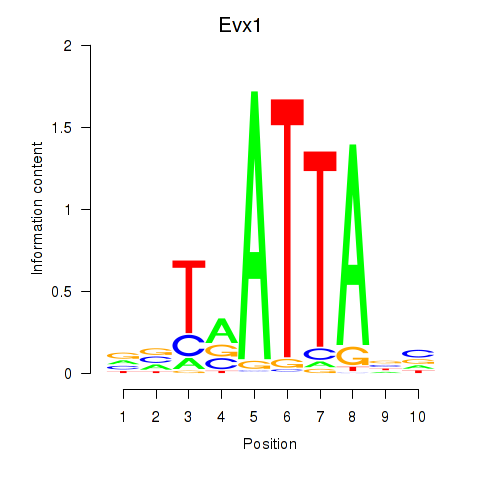
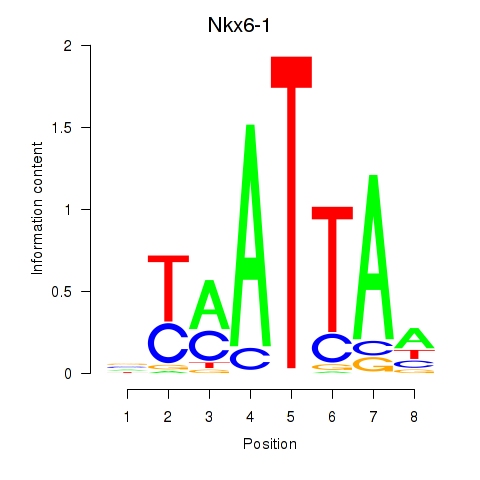
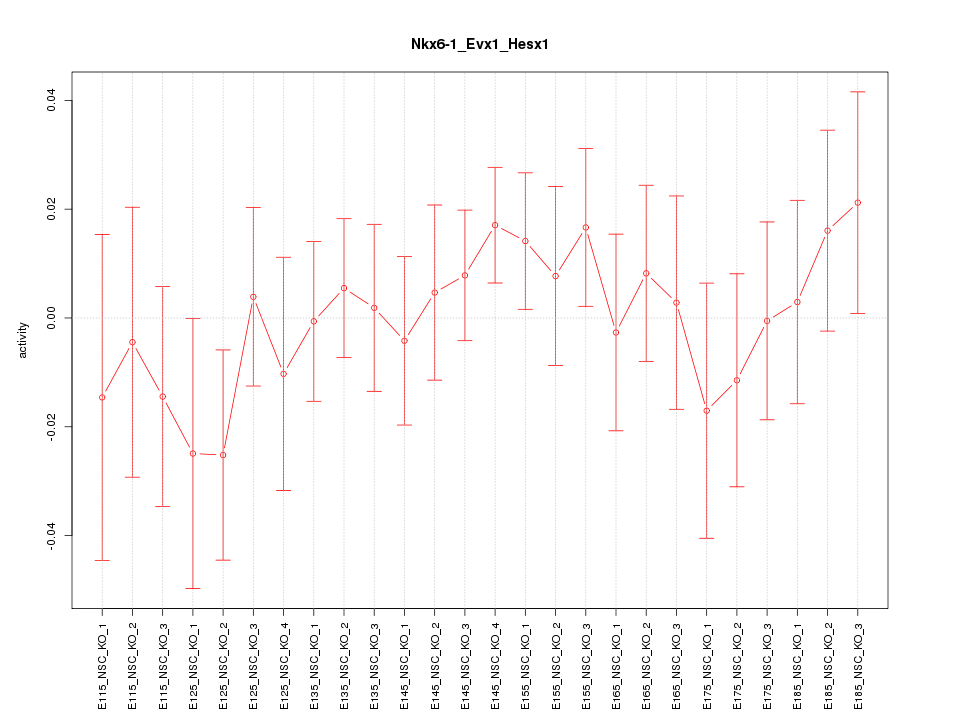
| 1.1 |
4.3 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.8 |
2.5 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 |
4.9 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.6 |
1.8 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.4 |
1.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.4 |
1.2 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.4 |
2.3 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 |
3.8 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 |
1.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.4 |
1.1 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.3 |
1.3 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 |
1.0 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 |
0.8 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 |
1.6 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
1.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
0.7 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 0.2 |
0.7 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 |
1.5 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.2 |
0.6 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.2 |
0.9 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.2 |
0.6 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 |
1.3 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
1.1 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 |
0.9 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.2 |
0.5 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.2 |
1.2 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
2.4 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.7 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.6 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.4 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.1 |
1.1 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 |
1.1 |
GO:0021830 |
interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.1 |
1.2 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.4 |
GO:0061738 |
abscission(GO:0009838) late endosomal microautophagy(GO:0061738) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.5 |
GO:0071205 |
protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.3 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.6 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
0.3 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
1.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.3 |
GO:0061055 |
myotome development(GO:0061055) |
| 0.1 |
0.8 |
GO:0002488 |
antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.1 |
0.8 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
0.4 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 |
3.1 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.2 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 |
1.2 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.4 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.1 |
0.3 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.5 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.2 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
1.5 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.1 |
0.6 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 |
1.7 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.3 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.3 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 |
0.4 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.4 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
1.2 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.1 |
0.3 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.1 |
4.3 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 |
0.1 |
GO:0097107 |
postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.3 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.2 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.3 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 |
0.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.3 |
GO:0032782 |
bile acid secretion(GO:0032782) |
| 0.1 |
0.2 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
1.0 |
GO:0046464 |
neutral lipid catabolic process(GO:0046461) acylglycerol catabolic process(GO:0046464) |
| 0.1 |
1.4 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.2 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 |
0.7 |
GO:0061050 |
regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 |
0.1 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.0 |
0.7 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.1 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.0 |
0.2 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.3 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0033762 |
response to glucagon(GO:0033762) response to mercury ion(GO:0046689) |
| 0.0 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.0 |
0.1 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 |
0.3 |
GO:0032264 |
IMP salvage(GO:0032264) |
| 0.0 |
0.1 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 |
0.2 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 |
0.2 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.1 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.0 |
0.1 |
GO:0042420 |
dopamine catabolic process(GO:0042420) |
| 0.0 |
0.7 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0008228 |
opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 |
0.9 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0046208 |
spermine catabolic process(GO:0046208) |
| 0.0 |
0.2 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.2 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
1.2 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.3 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.3 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.7 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.1 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.0 |
0.1 |
GO:0007494 |
midgut development(GO:0007494) |
| 0.0 |
0.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.3 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
0.4 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.1 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.0 |
0.4 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.2 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.0 |
0.2 |
GO:0046292 |
formaldehyde metabolic process(GO:0046292) |
| 0.0 |
1.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.5 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.1 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.7 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.8 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
1.0 |
GO:0007528 |
neuromuscular junction development(GO:0007528) |
| 0.0 |
0.6 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
2.4 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 |
0.4 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.1 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.0 |
0.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.4 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.0 |
GO:0045852 |
pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.3 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.3 |
GO:0009408 |
response to heat(GO:0009408) |
| 0.0 |
0.3 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
0.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.1 |
GO:0045602 |
negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 |
0.2 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.1 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
0.6 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.0 |
1.0 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.0 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
2.2 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
1.3 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.0 |
0.1 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.2 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.1 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.9 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.1 |
GO:0071286 |
cellular response to magnesium ion(GO:0071286) |
| 0.0 |
0.3 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
1.4 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.0 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.1 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.0 |
GO:0072362 |
regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 |
0.4 |
GO:0019915 |
lipid storage(GO:0019915) |
| 0.0 |
0.1 |
GO:1901898 |
regulation of cell communication by electrical coupling(GO:0010649) negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.0 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |