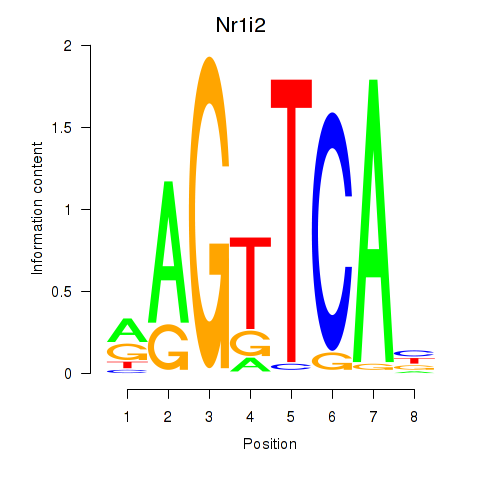
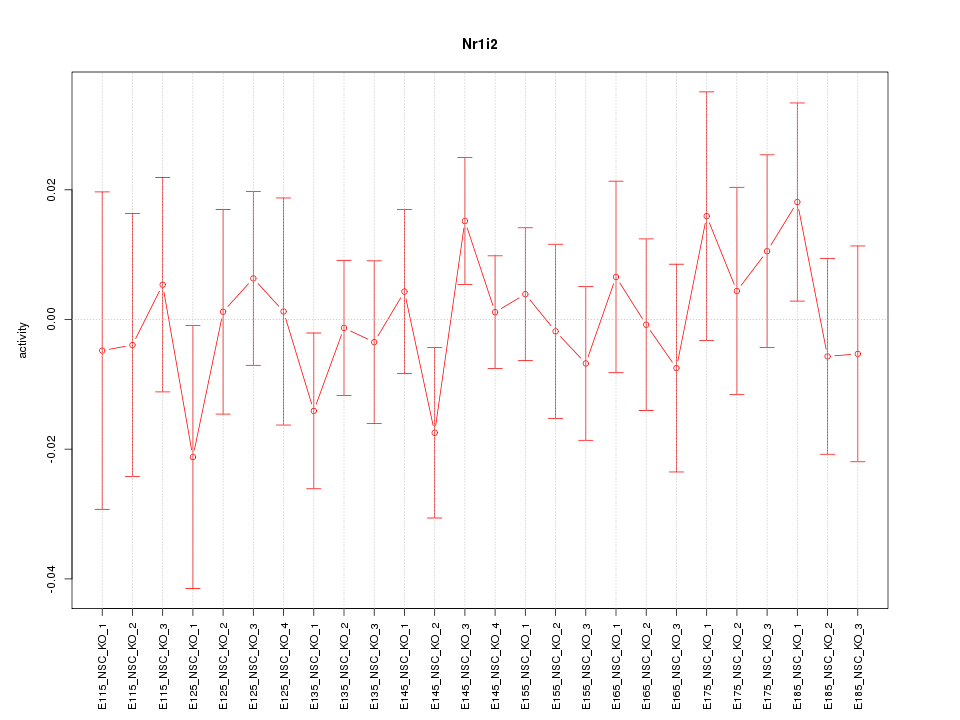
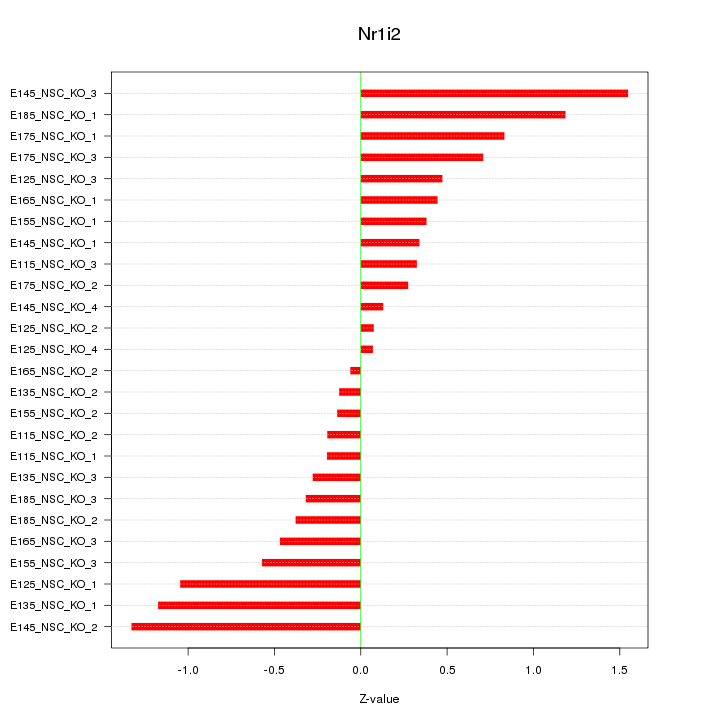
| 0.4 |
1.2 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.3 |
1.0 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 |
0.9 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.3 |
0.9 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.3 |
1.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 |
1.7 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 |
1.2 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.2 |
0.7 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.2 |
0.7 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 |
0.5 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.2 |
0.5 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.2 |
0.5 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
0.5 |
GO:1901731 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) positive regulation of platelet aggregation(GO:1901731) |
| 0.1 |
0.4 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.1 |
0.7 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.1 |
1.4 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
1.3 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 |
1.0 |
GO:0097460 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) |
| 0.1 |
0.6 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.6 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.9 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.7 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
1.1 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 |
0.3 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.5 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.1 |
0.4 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 |
0.9 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 |
0.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.1 |
0.5 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.2 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 |
0.4 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.7 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.1 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.1 |
0.4 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 |
0.4 |
GO:1901524 |
regulation of autophagosome maturation(GO:1901096) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.1 |
0.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
0.2 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
0.1 |
GO:0070601 |
centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 |
0.2 |
GO:0051695 |
actin filament uncapping(GO:0051695) |
| 0.1 |
0.2 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.8 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.1 |
0.2 |
GO:0042196 |
dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.1 |
0.3 |
GO:0042985 |
negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 |
0.3 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.1 |
0.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.0 |
0.1 |
GO:0042939 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.6 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.3 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.3 |
GO:2001023 |
regulation of response to drug(GO:2001023) |
| 0.0 |
0.1 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.2 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 |
0.1 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.0 |
1.0 |
GO:0006054 |
N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 |
0.9 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.2 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.0 |
0.1 |
GO:0060023 |
noradrenergic neuron differentiation(GO:0003357) soft palate development(GO:0060023) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.5 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.3 |
GO:0051036 |
regulation of endosome size(GO:0051036) |
| 0.0 |
0.9 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:1900157 |
regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 |
0.1 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 |
0.5 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.5 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.2 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 |
0.2 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.8 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.7 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.9 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.1 |
GO:1903903 |
striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 |
0.8 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 0.0 |
0.6 |
GO:0031958 |
corticosteroid receptor signaling pathway(GO:0031958) |
| 0.0 |
0.1 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 |
0.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.4 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.0 |
0.2 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 |
0.1 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.0 |
0.5 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.0 |
0.4 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.2 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.1 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 0.0 |
0.8 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.6 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.0 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.0 |
0.1 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.0 |
0.1 |
GO:0010572 |
positive regulation of platelet activation(GO:0010572) |
| 0.0 |
0.4 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.4 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.0 |
0.1 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.0 |
0.3 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.2 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.0 |
0.3 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
1.5 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.4 |
GO:0071357 |
type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 |
0.2 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.1 |
GO:0033602 |
negative regulation of dopamine secretion(GO:0033602) |
| 0.0 |
0.4 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.2 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.1 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.0 |
0.3 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.1 |
GO:0090611 |
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 |
0.2 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.2 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.0 |
0.6 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
2.0 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.5 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.3 |
GO:0071549 |
response to dexamethasone(GO:0071548) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 |
0.0 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) negative regulation of response to alcohol(GO:1901420) |
| 0.0 |
0.1 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.0 |
0.3 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0006659 |
phosphatidylserine biosynthetic process(GO:0006659) positive regulation of heat generation(GO:0031652) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 |
0.2 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 |
0.3 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 |
0.2 |
GO:0035999 |
tetrahydrofolate interconversion(GO:0035999) |
| 0.0 |
0.1 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 |
0.1 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.1 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.0 |
0.1 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 |
0.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.0 |
0.6 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.2 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.2 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.0 |
0.3 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.2 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |
| 0.0 |
0.3 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.3 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.5 |
GO:0001893 |
maternal placenta development(GO:0001893) |
| 0.0 |
0.1 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
0.3 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 |
0.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.1 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.0 |
0.0 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 |
0.2 |
GO:0030575 |
nuclear body organization(GO:0030575) |
| 0.0 |
0.1 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 |
0.4 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.1 |
GO:0019348 |
dolichol metabolic process(GO:0019348) |
| 0.0 |
0.1 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 |
0.3 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.1 |
GO:0046416 |
D-amino acid metabolic process(GO:0046416) |
| 0.0 |
0.3 |
GO:2000249 |
regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.2 |
GO:0033006 |
regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 |
0.2 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.1 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.0 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.0 |
0.2 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.3 |
GO:0022617 |
extracellular matrix disassembly(GO:0022617) |
| 0.0 |
0.0 |
GO:1902724 |
positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) positive regulation of metalloendopeptidase activity(GO:1904685) |
| 0.0 |
0.0 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 |
0.0 |
GO:0003211 |
cardiac ventricle formation(GO:0003211) |
| 0.0 |
0.1 |
GO:0019370 |
leukotriene biosynthetic process(GO:0019370) |
| 0.0 |
1.5 |
GO:0030041 |
actin filament polymerization(GO:0030041) |
| 0.0 |
0.4 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |