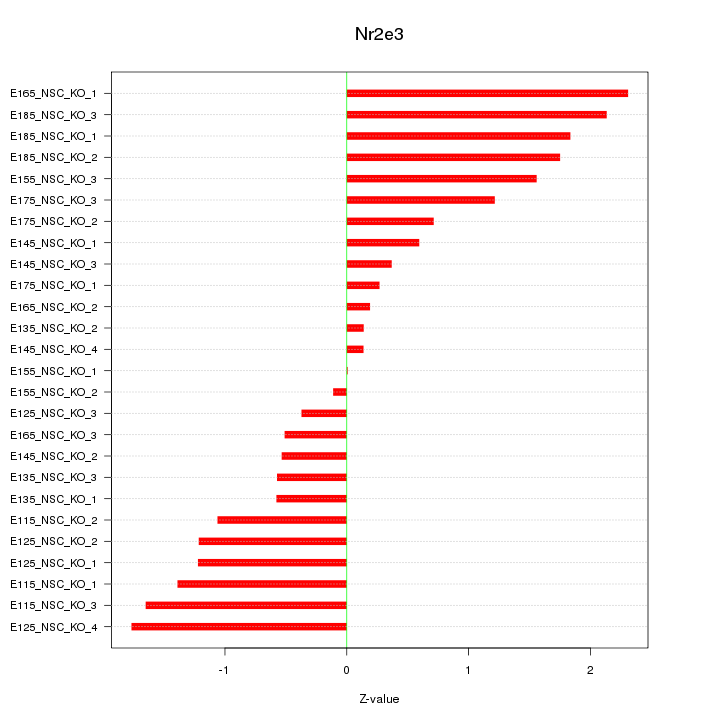
Motif ID: Nr2e3
Z-value: 1.154
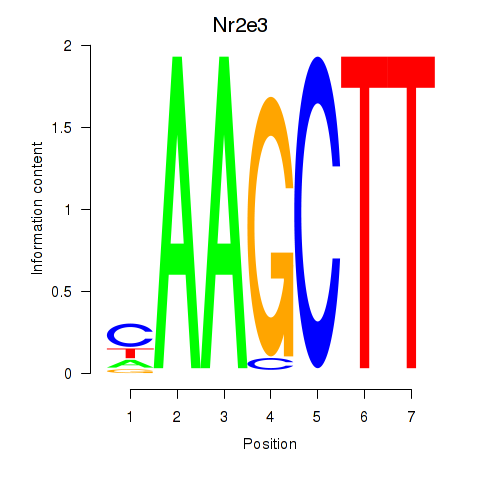
Transcription factors associated with Nr2e3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2e3 | ENSMUSG00000032292.2 | Nr2e3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.8 | 6.6 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.8 | 8.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.8 | 3.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.6 | 2.4 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.6 | 7.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 3.2 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.5 | 5.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.4 | 3.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.4 | 1.8 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.4 | 1.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.4 | 1.1 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.3 | 6.9 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.3 | 1.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 4.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 1.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 3.9 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 0.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.2 | 1.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 2.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 2.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.0 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.9 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 5.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 | 2.2 | GO:0090129 | regulation of synapse maturation(GO:0090128) positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.4 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.8 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 2.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.5 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 2.8 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.1 | 1.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 1.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 2.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.2 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.1 | 0.4 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 1.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 2.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.2 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 | 5.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 3.7 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 1.9 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.9 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 1.7 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.7 | 2.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.5 | 1.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 2.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 3.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 2.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 2.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.9 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 5.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 3.9 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 2.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 16.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 2.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 1.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 1.0 | GO:0042641 | actomyosin(GO:0042641) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.1 | 6.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 5.0 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.6 | 1.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.6 | 2.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 1.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.4 | 6.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.8 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 1.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 6.9 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.3 | 2.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 2.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 2.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 4.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 7.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 3.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 3.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 1.0 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.0 | 5.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.3 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.0 | 2.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 5.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.4 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 3.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.6 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |