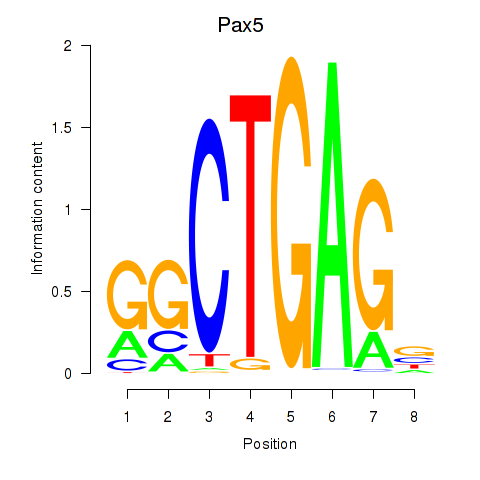
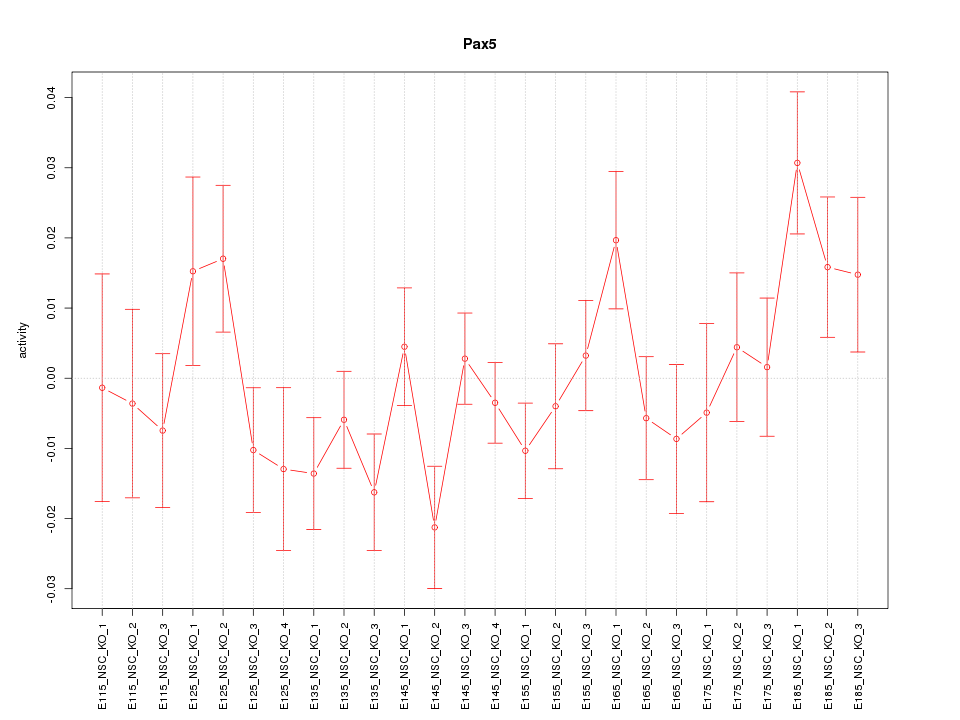
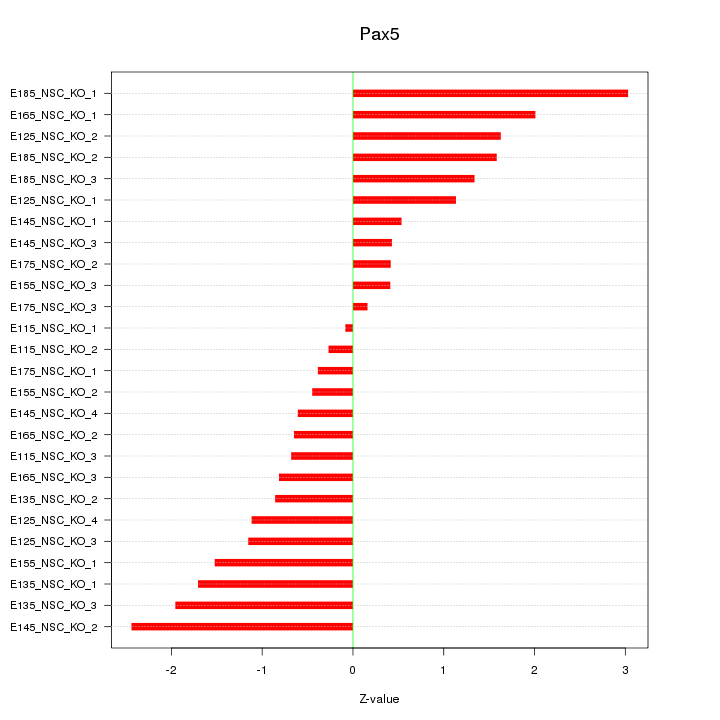
| 0.8 |
4.8 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 0.7 |
2.0 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.6 |
1.9 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.4 |
1.3 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.4 |
1.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.4 |
1.5 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.4 |
1.4 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.3 |
1.0 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.3 |
1.9 |
GO:0033600 |
negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.3 |
0.9 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 |
0.9 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 |
4.0 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.3 |
1.2 |
GO:2000427 |
positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 |
0.9 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.3 |
1.7 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 |
0.8 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 |
0.8 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.3 |
0.8 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.2 |
3.0 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 |
1.0 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.2 |
0.5 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 |
0.7 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 |
0.7 |
GO:0072076 |
nephrogenic mesenchyme development(GO:0072076) |
| 0.2 |
0.7 |
GO:0010751 |
regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.2 |
2.7 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.2 |
0.7 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 |
2.2 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.2 |
0.7 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 |
0.6 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
0.4 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 |
1.9 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 |
0.2 |
GO:0018307 |
enzyme active site formation(GO:0018307) |
| 0.2 |
2.8 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 |
1.0 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) |
| 0.2 |
0.6 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 |
0.9 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.2 |
0.7 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.2 |
3.9 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.2 |
1.4 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.2 |
0.8 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.2 |
0.8 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 |
0.5 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 |
0.5 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.2 |
0.6 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.2 |
0.5 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.1 |
0.6 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.1 |
0.4 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
0.1 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
0.9 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 |
0.4 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.1 |
1.0 |
GO:0061373 |
mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 |
0.5 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.5 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 |
0.5 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 |
0.7 |
GO:0050847 |
progesterone receptor signaling pathway(GO:0050847) |
| 0.1 |
0.4 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
0.5 |
GO:0060745 |
mammary gland branching involved in pregnancy(GO:0060745) |
| 0.1 |
0.6 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.1 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 0.1 |
0.4 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
0.5 |
GO:0002074 |
extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 |
0.4 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.1 |
0.8 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
0.4 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.1 |
1.0 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.1 |
0.5 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 |
0.5 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.2 |
GO:0046881 |
sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 |
0.3 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
0.4 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
0.6 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.1 |
0.3 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.1 |
0.3 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.5 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 |
0.4 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.2 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 |
0.3 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 |
1.6 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.1 |
0.7 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.1 |
2.7 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.1 |
2.0 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 |
0.4 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) regulation of membrane tubulation(GO:1903525) |
| 0.1 |
0.3 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.1 |
0.7 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
1.3 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 |
0.8 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.1 |
0.5 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 |
1.3 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
0.4 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.1 |
0.5 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
1.2 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 |
0.3 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.1 |
0.3 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 |
0.4 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
0.4 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
0.7 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.1 |
2.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.2 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 |
0.4 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.5 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.2 |
GO:0032916 |
transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.1 |
0.6 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 |
0.5 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.3 |
GO:0032229 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.4 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.3 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.1 |
0.4 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
1.2 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.1 |
0.6 |
GO:0048012 |
hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 |
0.4 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
1.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 |
0.4 |
GO:0034372 |
very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 |
0.4 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 0.1 |
0.3 |
GO:0001923 |
B-1 B cell differentiation(GO:0001923) |
| 0.1 |
0.1 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
0.3 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.1 |
0.1 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.1 |
1.9 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.1 |
0.7 |
GO:0097421 |
liver regeneration(GO:0097421) |
| 0.1 |
0.5 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
0.2 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.1 |
0.1 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.1 |
0.7 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.3 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.1 |
0.3 |
GO:0021940 |
positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 |
0.3 |
GO:1903546 |
microtubule anchoring at centrosome(GO:0034454) negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 |
0.8 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.6 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.4 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
0.4 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 |
0.3 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
0.5 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.1 |
1.6 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
0.2 |
GO:0048245 |
eosinophil chemotaxis(GO:0048245) |
| 0.1 |
0.2 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 |
0.7 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 |
0.1 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.1 |
0.4 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.2 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
0.4 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.3 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
1.1 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.1 |
0.2 |
GO:2000612 |
regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072039) negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072040) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:1901145) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of somatic stem cell population maintenance(GO:1904672) negative regulation of somatic stem cell population maintenance(GO:1904673) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 |
0.1 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 |
0.3 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
0.2 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.3 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 |
0.2 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.7 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.2 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.1 |
0.7 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.5 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 |
0.2 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
0.6 |
GO:0045332 |
phospholipid translocation(GO:0045332) |
| 0.1 |
0.5 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
0.9 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 |
0.6 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
1.1 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.4 |
GO:2000781 |
positive regulation of double-strand break repair(GO:2000781) |
| 0.0 |
0.9 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.4 |
GO:0033689 |
negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.0 |
1.0 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.0 |
0.6 |
GO:0060394 |
negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 |
0.3 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 |
0.2 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
3.2 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 |
0.8 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 |
0.1 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.0 |
0.4 |
GO:0019673 |
dolichol metabolic process(GO:0019348) GDP-mannose metabolic process(GO:0019673) |
| 0.0 |
0.9 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:1900242 |
regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 |
0.1 |
GO:0090069 |
regulation of ribosome biogenesis(GO:0090069) |
| 0.0 |
0.3 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.4 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.3 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.9 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.4 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 |
0.4 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 |
0.6 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.0 |
0.1 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 |
0.4 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.4 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.6 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.5 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.2 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.2 |
GO:0045347 |
negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 |
0.2 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 |
0.2 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 |
0.3 |
GO:0061088 |
sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 |
0.2 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.2 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.1 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 |
1.6 |
GO:0046326 |
positive regulation of glucose import(GO:0046326) |
| 0.0 |
0.6 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
0.2 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.6 |
GO:0031103 |
axon regeneration(GO:0031103) |
| 0.0 |
0.2 |
GO:0021514 |
ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 |
0.4 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.3 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.1 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.0 |
0.2 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.8 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.3 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.0 |
1.3 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 |
1.0 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.6 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
2.5 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
1.4 |
GO:0048278 |
vesicle docking(GO:0048278) |
| 0.0 |
0.5 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 |
0.7 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.1 |
GO:0016540 |
protein autoprocessing(GO:0016540) |
| 0.0 |
0.5 |
GO:0030204 |
chondroitin sulfate metabolic process(GO:0030204) |
| 0.0 |
0.3 |
GO:0042403 |
thyroid hormone metabolic process(GO:0042403) |
| 0.0 |
0.1 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 |
0.4 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.1 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.0 |
1.3 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.5 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.0 |
0.3 |
GO:2000300 |
regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 |
0.8 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.7 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.3 |
GO:0044539 |
long-chain fatty acid import(GO:0044539) |
| 0.0 |
0.2 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 |
0.5 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.1 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
0.1 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 |
0.1 |
GO:0048669 |
collateral sprouting in absence of injury(GO:0048669) |
| 0.0 |
0.3 |
GO:0010960 |
magnesium ion homeostasis(GO:0010960) |
| 0.0 |
1.0 |
GO:0021510 |
spinal cord development(GO:0021510) |
| 0.0 |
0.8 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.4 |
GO:1903779 |
regulation of cardiac conduction(GO:1903779) |
| 0.0 |
0.0 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.0 |
0.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.3 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.3 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.8 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.0 |
0.3 |
GO:0019886 |
antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 |
0.2 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.8 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.7 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.0 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 |
1.6 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.1 |
GO:0071440 |
regulation of histone H3-K14 acetylation(GO:0071440) |
| 0.0 |
0.3 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.0 |
0.3 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.5 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.2 |
GO:0003148 |
outflow tract septum morphogenesis(GO:0003148) |
| 0.0 |
0.2 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.0 |
0.0 |
GO:0090188 |
negative regulation of pancreatic juice secretion(GO:0090188) |
| 0.0 |
0.5 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.1 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 |
0.2 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.0 |
0.1 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.0 |
0.1 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.3 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.6 |
GO:0017145 |
stem cell division(GO:0017145) |
| 0.0 |
0.2 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
0.3 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 |
0.1 |
GO:0015868 |
purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.2 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.1 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 |
0.3 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.1 |
GO:0036151 |
negative regulation of fatty acid beta-oxidation(GO:0031999) phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 |
0.1 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.0 |
0.1 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.1 |
GO:0032695 |
negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 |
0.4 |
GO:0045453 |
bone resorption(GO:0045453) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.1 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.0 |
0.3 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.4 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
0.2 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.0 |
0.4 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.0 |
0.3 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.5 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
0.4 |
GO:0032007 |
negative regulation of TOR signaling(GO:0032007) |
| 0.0 |
0.0 |
GO:0097484 |
dendrite extension(GO:0097484) |
| 0.0 |
0.2 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.0 |
0.4 |
GO:0006111 |
regulation of gluconeogenesis(GO:0006111) |
| 0.0 |
0.2 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.0 |
0.2 |
GO:0014044 |
Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.2 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0071569 |
protein ufmylation(GO:0071569) |
| 0.0 |
0.1 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 |
0.8 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.3 |
GO:0010812 |
negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 |
0.1 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
0.2 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.1 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
0.2 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
0.1 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.1 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 |
0.1 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.0 |
0.5 |
GO:0007338 |
single fertilization(GO:0007338) |
| 0.0 |
0.3 |
GO:0009395 |
phospholipid catabolic process(GO:0009395) |
| 0.0 |
0.1 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.1 |
GO:0072643 |
interferon-gamma secretion(GO:0072643) |