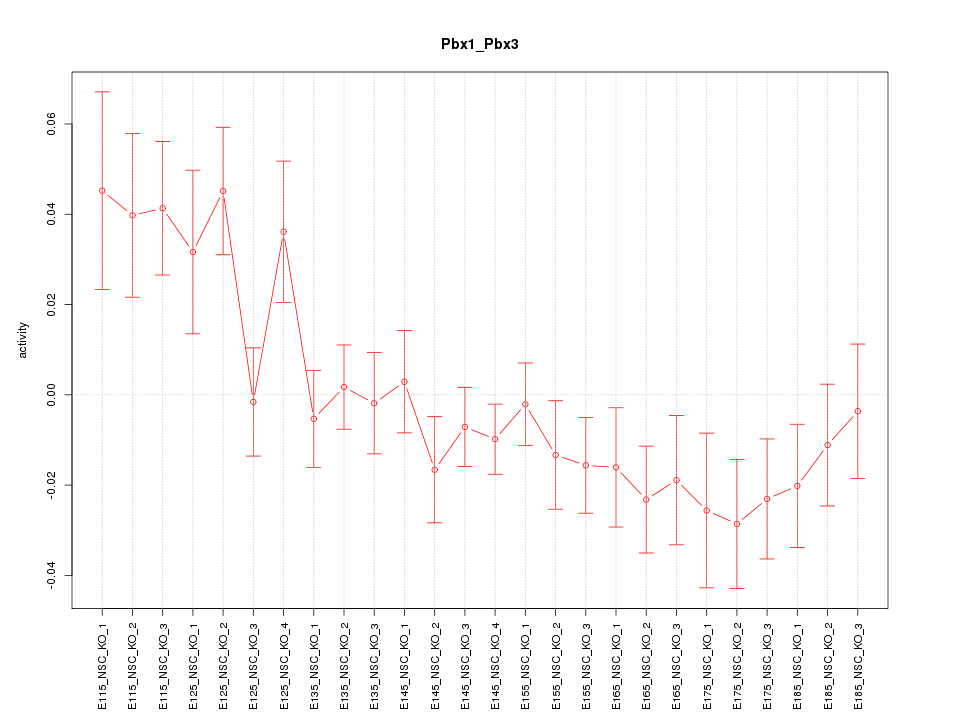
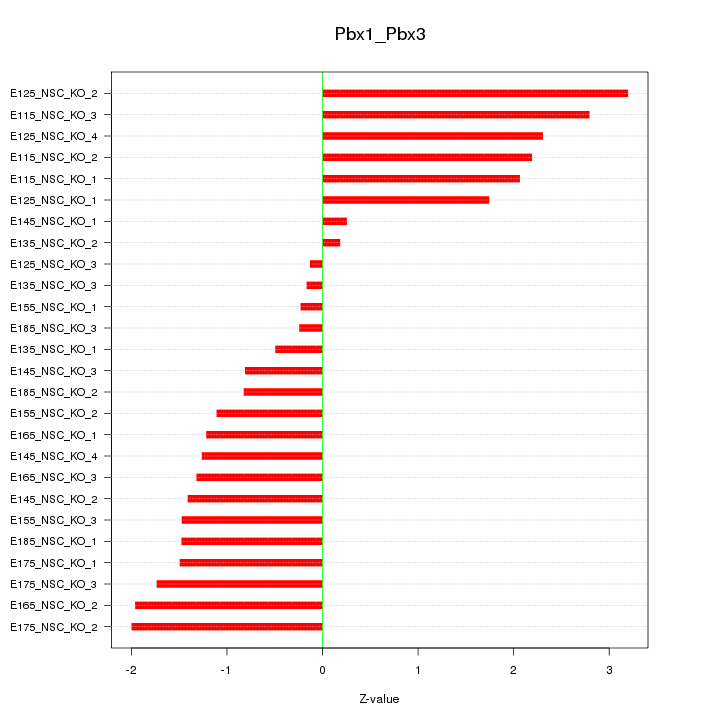
Motif ID: Pbx1_Pbx3
Z-value: 1.554

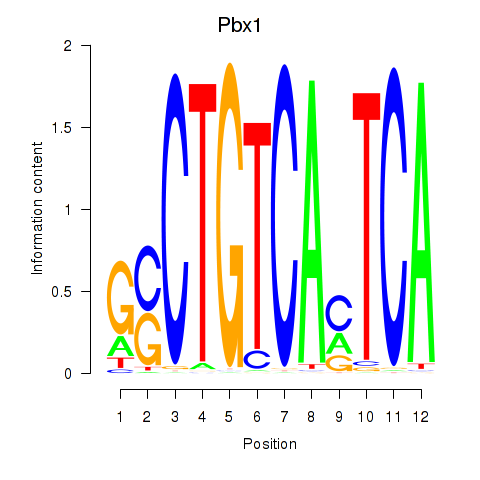
Transcription factors associated with Pbx1_Pbx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pbx1 | ENSMUSG00000052534.9 | Pbx1 |
| Pbx3 | ENSMUSG00000038718.9 | Pbx3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pbx1 | mm10_v2_chr1_-_168431502_168431513 | -0.71 | 5.5e-05 | Click! |
| Pbx3 | mm10_v2_chr2_-_34372004_34372044 | 0.68 | 1.3e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0097402 | neuroblast migration(GO:0097402) |
| 3.8 | 30.5 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 3.2 | 9.6 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) positive regulation of transforming growth factor beta3 production(GO:0032916) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 2.5 | 7.6 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 2.1 | 8.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.8 | 9.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 1.5 | 4.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.2 | 3.6 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.2 | 5.8 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 1.1 | 3.3 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.1 | 6.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.0 | 3.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.9 | 2.8 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.9 | 3.8 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.9 | 4.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.8 | 2.3 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.7 | 2.9 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.6 | 1.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.6 | 1.8 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.6 | 2.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.6 | 1.1 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.6 | 1.7 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.5 | 4.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 12.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.5 | 8.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.5 | 2.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 2.0 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.5 | 3.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.5 | 2.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 1.3 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.4 | 1.3 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 | 4.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 2.5 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.4 | 1.7 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.4 | 1.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 6.6 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.4 | 3.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.4 | 1.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.3 | 2.0 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 6.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.3 | 1.8 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.3 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 1.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.3 | 1.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.3 | 0.8 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.3 | 0.8 | GO:0071692 | sequestering of BMP in extracellular matrix(GO:0035582) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.2 | 3.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 4.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 2.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 0.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.2 | 2.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.2 | 0.4 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 3.6 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 2.6 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 2.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 2.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 1.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 0.7 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.5 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.2 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.3 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.2 | 0.5 | GO:0060672 | negative regulation of keratinocyte differentiation(GO:0045617) epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 3.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 1.6 | GO:0030388 | fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 1.5 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 1.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.2 | 2.1 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 0.9 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 1.8 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.3 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.1 | 1.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 1.5 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.5 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.1 | 0.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.5 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 5.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.9 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.1 | 1.6 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 1.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.2 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.9 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 0.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 4.4 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 0.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) positive regulation of histone H3-K9 acetylation(GO:2000617) |
| 0.0 | 0.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.7 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.5 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 4.7 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 4.7 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.9 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 4.1 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 2.1 | GO:0001942 | hair follicle development(GO:0001942) molting cycle process(GO:0022404) hair cycle process(GO:0022405) skin epidermis development(GO:0098773) |
| 0.0 | 1.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 1.2 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.2 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 1.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 1.4 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.4 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 1.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.8 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.3 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 2.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 30.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 1.9 | 9.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 1.6 | 4.7 | GO:0000801 | central element(GO:0000801) |
| 1.2 | 11.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.8 | 4.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.6 | 1.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 2.8 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 3.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.4 | 2.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.4 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.4 | 2.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 1.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 1.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 1.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 2.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 7.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 2.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 4.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 4.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.8 | GO:0001527 | microfibril(GO:0001527) |
| 0.1 | 22.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.4 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.6 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 6.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.7 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.6 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 2.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 1.5 | 4.5 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.4 | 4.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.3 | 31.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.8 | 15.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.8 | 3.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.7 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 0.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.5 | 5.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 1.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.4 | 2.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.4 | 6.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 1.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 2.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 4.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.3 | 1.4 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.3 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 2.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 4.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 2.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.3 | 1.8 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.3 | 3.9 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.3 | 3.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 2.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.0 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 1.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.2 | 0.6 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 21.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.2 | 1.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 0.7 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.5 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.2 | 2.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 0.5 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.5 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 3.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 3.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.1 | 11.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 44.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 9.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 9.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.6 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 1.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 4.1 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 4.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 1.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 2.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 3.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.4 | GO:0001094 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 5.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.5 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.3 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.9 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.9 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.2 | GO:0051015 | actin filament binding(GO:0051015) |