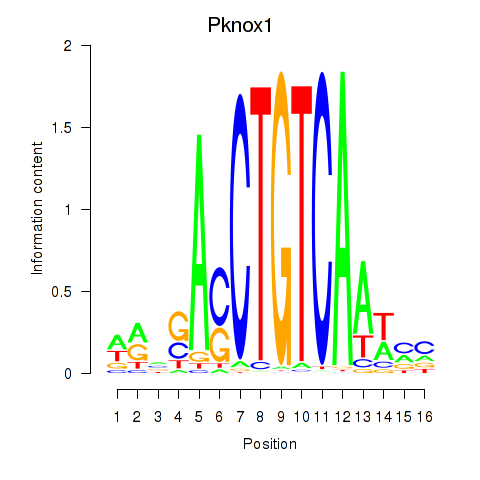
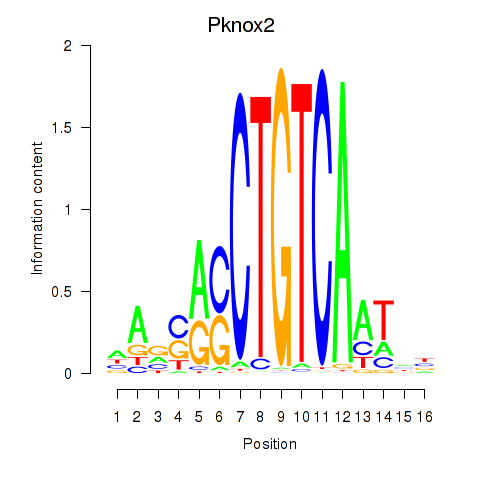
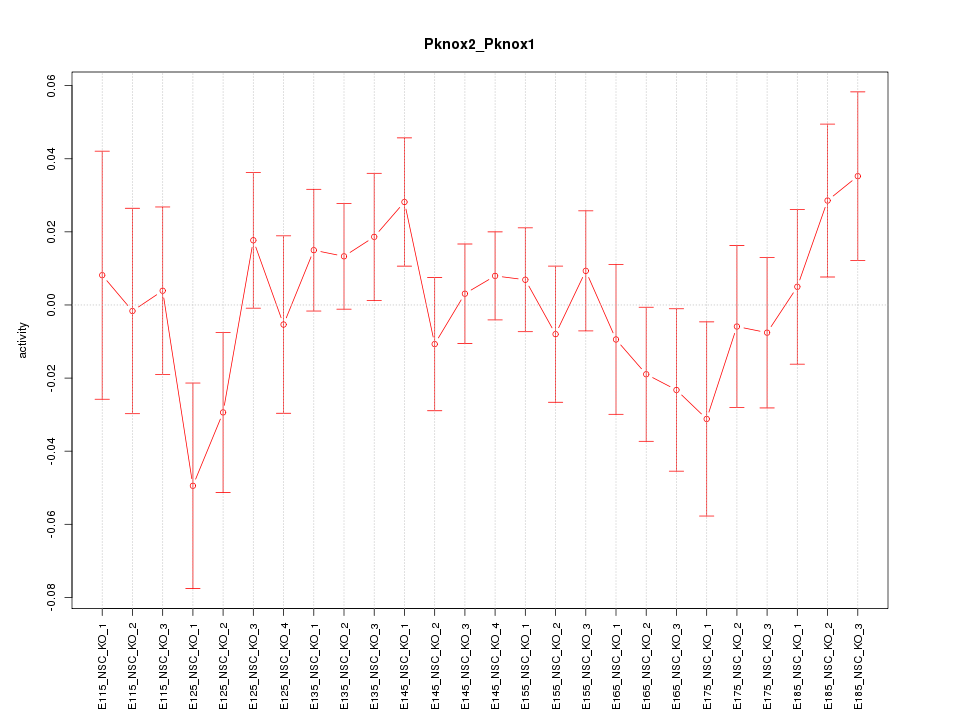
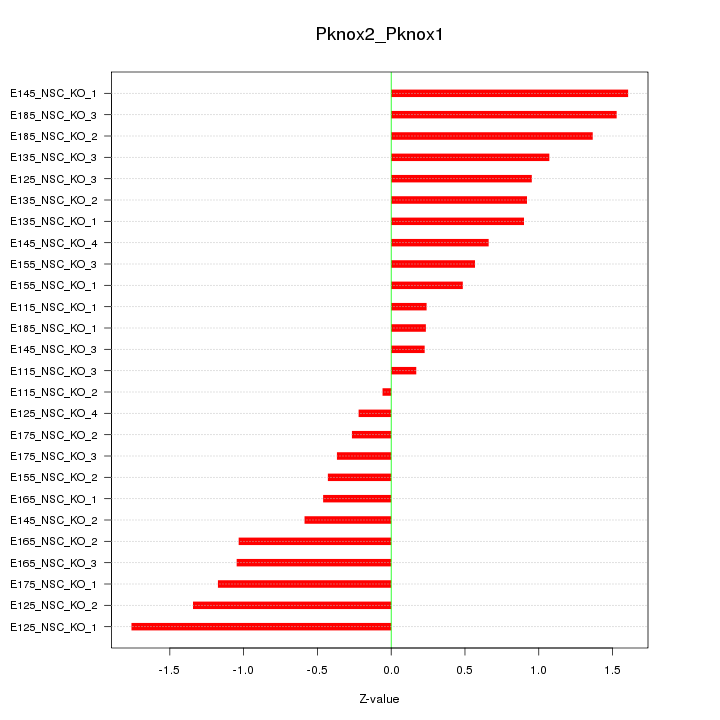
| 0.9 |
2.7 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.4 |
1.5 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 |
1.6 |
GO:0021905 |
pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.3 |
2.9 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 |
1.7 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
3.4 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 |
1.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
0.6 |
GO:1901228 |
regulation of osteoclast proliferation(GO:0090289) negative regulation of bone mineralization involved in bone maturation(GO:1900158) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.2 |
0.6 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.2 |
1.3 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 |
0.5 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.2 |
1.8 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
1.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.6 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 |
1.6 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 |
0.7 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.7 |
GO:0006307 |
DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 |
0.7 |
GO:1905206 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 |
0.3 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
0.6 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 |
0.6 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.1 |
0.4 |
GO:2000325 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.1 |
0.6 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.5 |
GO:0051461 |
positive regulation of heat generation(GO:0031652) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.1 |
1.0 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.6 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.2 |
GO:2000813 |
actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 |
0.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.3 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.2 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) |
| 0.1 |
2.5 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
1.4 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 |
0.4 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.3 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.0 |
0.5 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.3 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.6 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.0 |
0.3 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.6 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.0 |
0.1 |
GO:0090202 |
transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 |
0.2 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 |
0.5 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.0 |
1.0 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.6 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.3 |
GO:0006972 |
hyperosmotic response(GO:0006972) activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.8 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.1 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 |
0.4 |
GO:0007099 |
centriole replication(GO:0007099) |
| 0.0 |
1.1 |
GO:0007187 |
G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 |
0.0 |
GO:1901896 |
positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 |
0.1 |
GO:0035510 |
DNA dealkylation(GO:0035510) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |