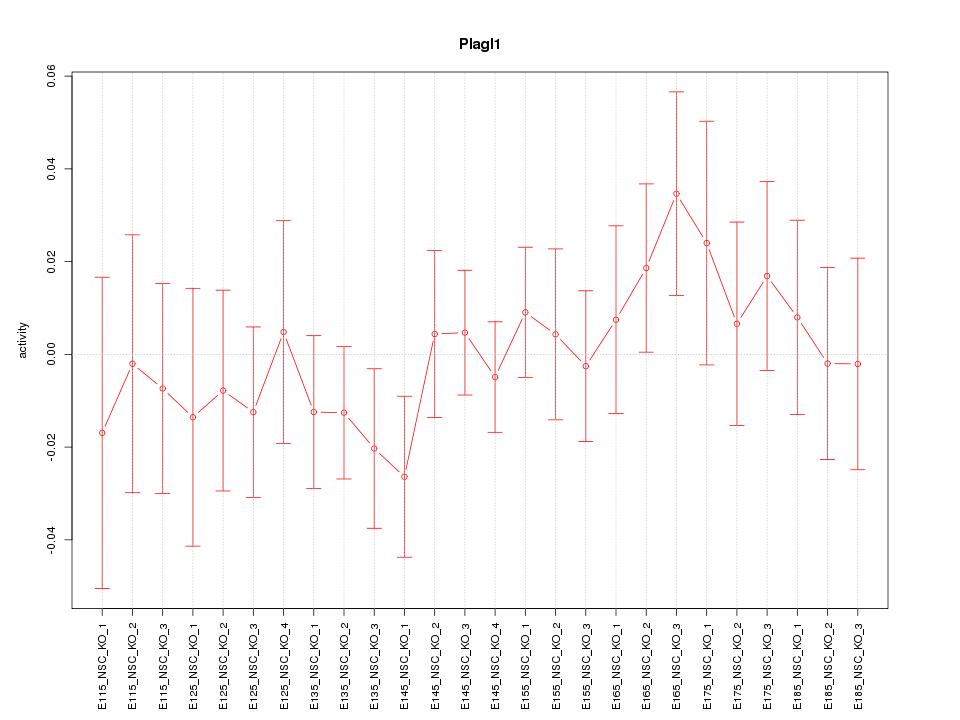
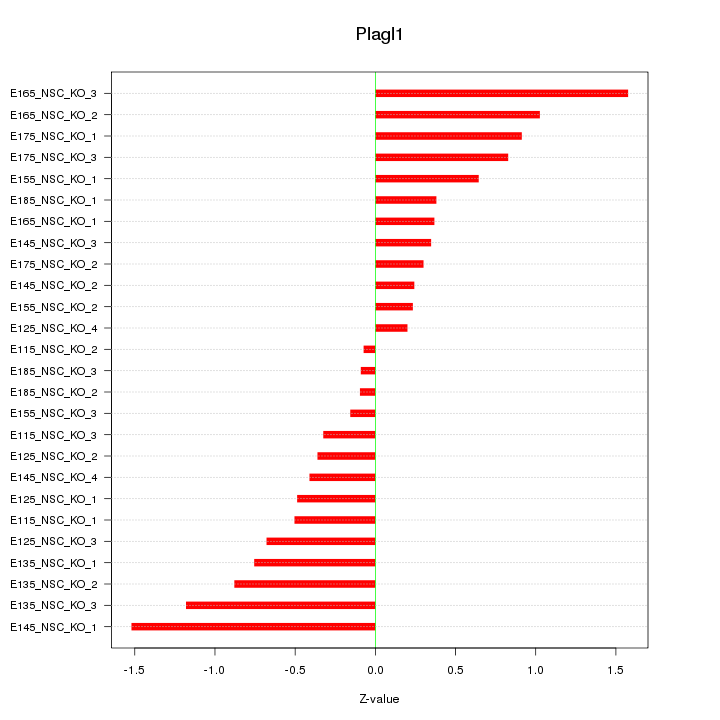
Motif ID: Plagl1
Z-value: 0.695

Transcription factors associated with Plagl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Plagl1 | ENSMUSG00000019817.12 | Plagl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | mm10_v2_chr10_+_13090788_13090843 | -0.82 | 2.5e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.5 | 2.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.5 | 4.6 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.3 | 0.9 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.7 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 | 0.9 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 1.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 1.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 2.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 0.5 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.9 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 1.8 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 1.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 4.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.9 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.7 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.1 | 0.6 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.9 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.6 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.0 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.9 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.6 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 2.6 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.4 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 1.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.7 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) eye pigmentation(GO:0048069) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.9 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.2 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.8 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 4.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.3 | 1.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.3 | 0.9 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.9 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.2 | 2.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 4.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.3 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 2.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.9 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.3 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |