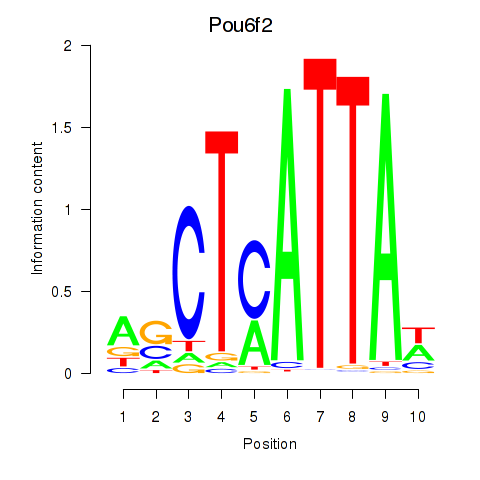
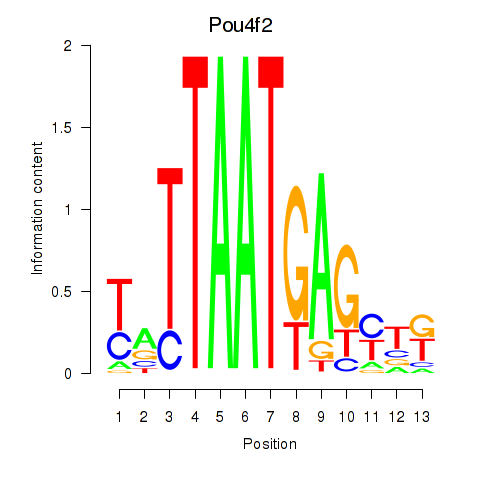
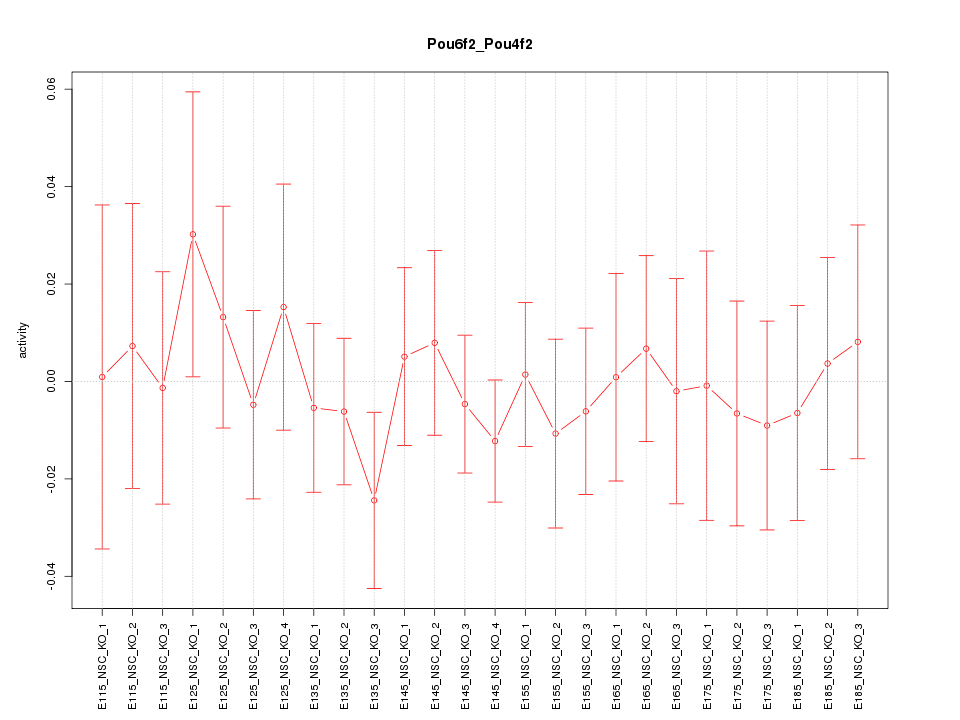
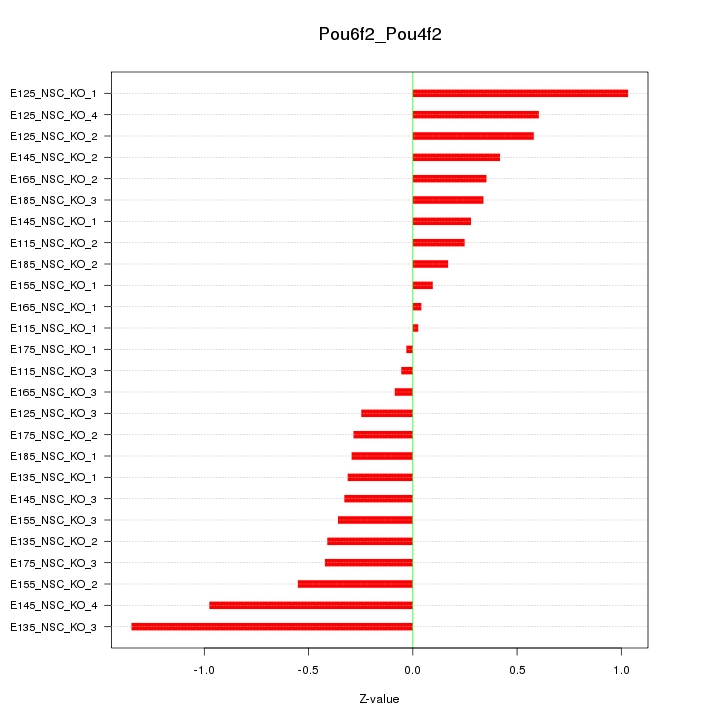
| 0.7 |
2.0 |
GO:0072204 |
cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.4 |
1.3 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.4 |
5.2 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 |
1.2 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.3 |
0.9 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.3 |
2.2 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 |
0.6 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 |
2.0 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.3 |
GO:0090324 |
negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.1 |
0.3 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.2 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 |
0.6 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.1 |
0.6 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.4 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.4 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.6 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.9 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.1 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.0 |
0.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.1 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 |
0.2 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.0 |
0.3 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.1 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.0 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.0 |
0.1 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.3 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.3 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |