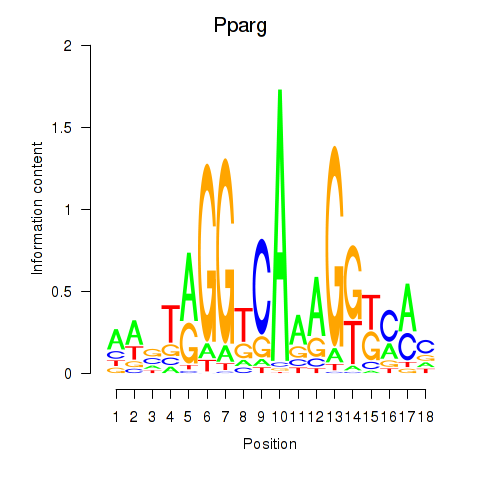
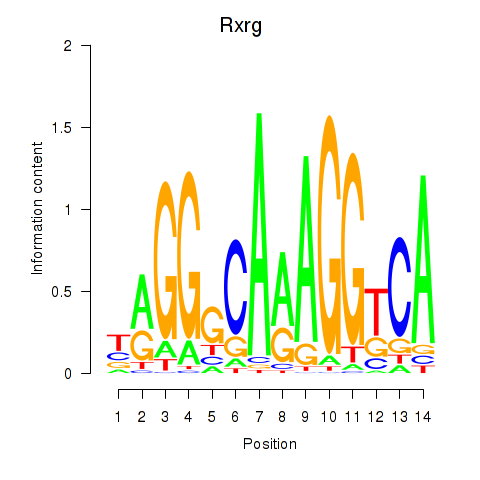
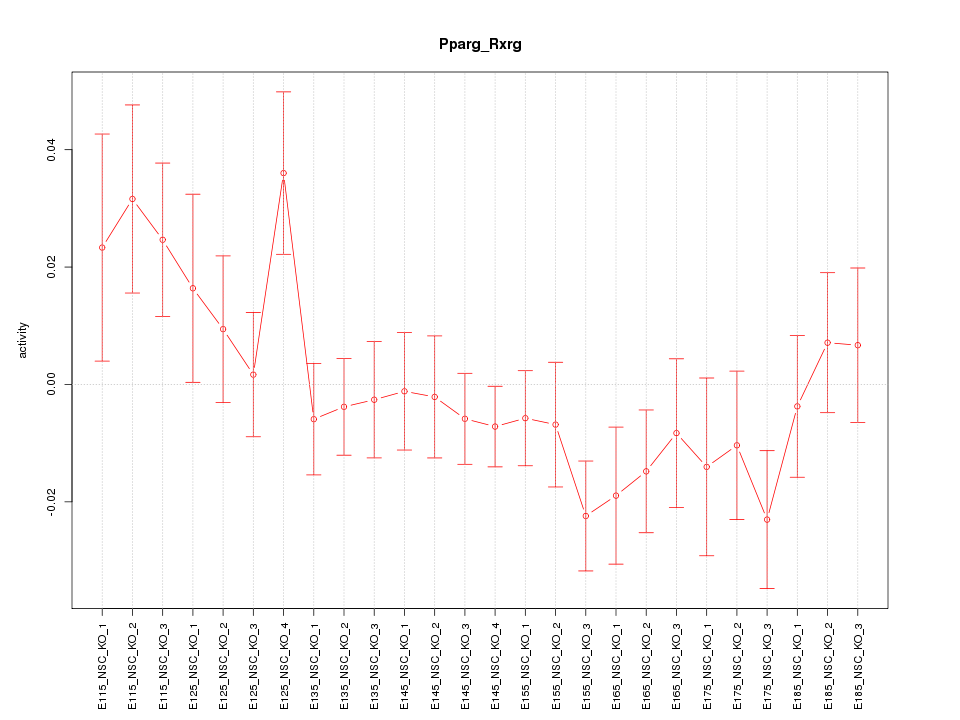
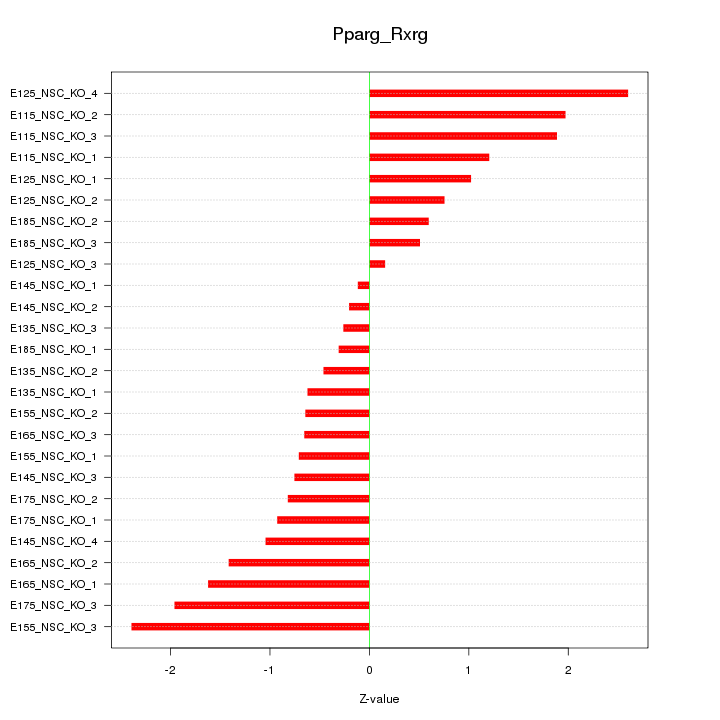
| 1.7 |
5.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 1.4 |
4.3 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.1 |
5.5 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.9 |
2.7 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.9 |
5.3 |
GO:0098700 |
aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.9 |
4.4 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.9 |
3.4 |
GO:0032901 |
positive regulation of neurotrophin production(GO:0032901) |
| 0.9 |
2.6 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.8 |
2.4 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.8 |
2.4 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.8 |
2.3 |
GO:0014028 |
notochord formation(GO:0014028) |
| 0.7 |
5.9 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.7 |
2.2 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.7 |
2.0 |
GO:1900211 |
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.7 |
4.7 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.6 |
1.9 |
GO:0006578 |
amino-acid betaine biosynthetic process(GO:0006578) |
| 0.6 |
6.0 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.6 |
3.6 |
GO:0006287 |
base-excision repair, gap-filling(GO:0006287) |
| 0.6 |
1.7 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.6 |
1.7 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.5 |
1.5 |
GO:0046381 |
CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 |
2.0 |
GO:0048682 |
axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.5 |
2.3 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.5 |
1.4 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.5 |
2.3 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.5 |
3.6 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.4 |
1.3 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.4 |
1.7 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 |
1.6 |
GO:1902302 |
regulation of potassium ion export(GO:1902302) |
| 0.4 |
1.2 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.4 |
2.0 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.4 |
1.8 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.4 |
1.8 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.3 |
1.7 |
GO:0015705 |
iodide transport(GO:0015705) |
| 0.3 |
2.4 |
GO:0071294 |
cellular response to zinc ion(GO:0071294) |
| 0.3 |
1.0 |
GO:0090274 |
positive regulation of somatostatin secretion(GO:0090274) |
| 0.3 |
2.0 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 |
0.6 |
GO:0006145 |
purine nucleobase catabolic process(GO:0006145) |
| 0.3 |
1.0 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.3 |
4.0 |
GO:0006000 |
fructose metabolic process(GO:0006000) |
| 0.3 |
2.2 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.3 |
5.5 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 |
1.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.3 |
0.9 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.3 |
5.9 |
GO:0001522 |
pseudouridine synthesis(GO:0001522) |
| 0.3 |
0.5 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 |
1.3 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 |
1.1 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 |
1.8 |
GO:2001199 |
negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.3 |
0.3 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 |
1.0 |
GO:0010890 |
positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.2 |
1.0 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
0.7 |
GO:0015920 |
lipopolysaccharide transport(GO:0015920) |
| 0.2 |
1.2 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.2 |
1.6 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 |
2.0 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 |
1.1 |
GO:0051593 |
response to folic acid(GO:0051593) |
| 0.2 |
0.6 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 |
1.9 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) |
| 0.2 |
0.6 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 |
0.4 |
GO:0002636 |
positive regulation of germinal center formation(GO:0002636) |
| 0.2 |
1.0 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.2 |
0.6 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.2 |
1.4 |
GO:0006703 |
estrogen biosynthetic process(GO:0006703) |
| 0.2 |
0.6 |
GO:0097461 |
ferric iron import(GO:0033216) ferric iron import into cell(GO:0097461) |
| 0.2 |
1.0 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.2 |
0.6 |
GO:0060282 |
positive regulation of chronic inflammatory response(GO:0002678) positive regulation of oocyte development(GO:0060282) |
| 0.2 |
2.5 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.2 |
0.4 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
0.6 |
GO:1902109 |
negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 |
1.3 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.2 |
0.6 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 |
0.6 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.2 |
1.1 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
0.7 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 |
0.5 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 |
0.5 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.2 |
1.6 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.2 |
1.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 |
0.9 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.2 |
1.6 |
GO:0046459 |
short-chain fatty acid metabolic process(GO:0046459) |
| 0.2 |
0.7 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.2 |
0.7 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.2 |
1.1 |
GO:0033567 |
DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 |
2.9 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.2 |
0.6 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.2 |
0.8 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.2 |
1.7 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
0.4 |
GO:0060010 |
Sertoli cell fate commitment(GO:0060010) |
| 0.1 |
0.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.7 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
5.5 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 |
0.9 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.1 |
2.4 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
1.9 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 |
0.4 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
1.6 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.4 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.1 |
0.7 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.1 |
1.3 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
0.4 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.1 |
0.8 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 |
0.7 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 |
0.7 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 |
0.4 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 |
0.4 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.7 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.1 |
0.7 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.1 |
0.8 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
1.2 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 |
0.3 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.1 |
0.4 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 |
0.5 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.6 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
1.2 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 |
0.6 |
GO:0046668 |
regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
1.0 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.8 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 |
0.8 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.1 |
1.3 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.7 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
0.2 |
GO:0036337 |
Fas signaling pathway(GO:0036337) |
| 0.1 |
1.0 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.5 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 |
0.3 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.6 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.1 |
0.5 |
GO:0060279 |
positive regulation of ovulation(GO:0060279) |
| 0.1 |
0.4 |
GO:0006438 |
valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 |
1.9 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
1.4 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.1 |
0.6 |
GO:0042256 |
mature ribosome assembly(GO:0042256) |
| 0.1 |
1.8 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.1 |
0.6 |
GO:0060018 |
astrocyte fate commitment(GO:0060018) |
| 0.1 |
1.2 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.3 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
0.3 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 |
0.7 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.1 |
0.1 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.1 |
0.3 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.4 |
GO:0071475 |
cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.1 |
0.3 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 |
0.3 |
GO:0030497 |
fatty acid elongation(GO:0030497) |
| 0.1 |
1.8 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 |
0.6 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 |
0.1 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 |
0.2 |
GO:0035771 |
interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 |
0.4 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
3.1 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
5.5 |
GO:0006446 |
regulation of translational initiation(GO:0006446) |
| 0.1 |
0.3 |
GO:0003365 |
establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 |
0.9 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
0.5 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.1 |
2.4 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.6 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
2.3 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.2 |
GO:0048611 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.4 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
0.2 |
GO:0006987 |
activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 |
0.2 |
GO:0098915 |
membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 |
1.0 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 |
0.3 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 |
0.9 |
GO:0046784 |
viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 |
0.3 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.1 |
2.9 |
GO:0007588 |
excretion(GO:0007588) |
| 0.1 |
0.8 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
0.6 |
GO:0042136 |
neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 |
0.5 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.5 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.1 |
0.5 |
GO:0051004 |
regulation of lipoprotein lipase activity(GO:0051004) |
| 0.1 |
2.6 |
GO:0045879 |
negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 |
0.4 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.6 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.6 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
1.5 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.1 |
0.8 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 |
0.6 |
GO:0051639 |
actin filament network formation(GO:0051639) |
| 0.1 |
0.6 |
GO:1904668 |
positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 |
0.8 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.4 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.1 |
1.0 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.1 |
3.8 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.1 |
1.4 |
GO:0048368 |
lateral mesoderm development(GO:0048368) |
| 0.1 |
0.2 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
0.3 |
GO:0046602 |
mitotic centrosome separation(GO:0007100) regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 |
0.6 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 |
0.5 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
0.5 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 |
0.6 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 |
0.6 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 |
1.0 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.1 |
0.6 |
GO:0001773 |
myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.5 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 |
0.2 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 |
0.4 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.3 |
GO:0072488 |
nitrogen utilization(GO:0019740) ammonium transmembrane transport(GO:0072488) |
| 0.1 |
0.4 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 |
0.6 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
0.5 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.1 |
0.2 |
GO:0002072 |
optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 |
0.2 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.1 |
GO:0003169 |
coronary vein morphogenesis(GO:0003169) |
| 0.1 |
0.6 |
GO:1902187 |
negative regulation of viral release from host cell(GO:1902187) |
| 0.1 |
0.2 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
0.4 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.3 |
GO:2000096 |
non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) convergent extension involved in axis elongation(GO:0060028) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.2 |
GO:0006290 |
pyrimidine dimer repair(GO:0006290) |
| 0.1 |
0.2 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.4 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.1 |
0.6 |
GO:0006824 |
cobalt ion transport(GO:0006824) |
| 0.1 |
1.0 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.5 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
0.3 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.0 |
1.7 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.4 |
GO:0006450 |
regulation of translational fidelity(GO:0006450) |
| 0.0 |
0.2 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.1 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 |
0.2 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 |
0.2 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.8 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.2 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.0 |
0.4 |
GO:0000467 |
exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 |
0.1 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 |
0.3 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.0 |
1.2 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
1.2 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.0 |
0.4 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.4 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.4 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.1 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 |
0.4 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 |
0.3 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
1.2 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.0 |
0.5 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 |
0.1 |
GO:0035461 |
vitamin transmembrane transport(GO:0035461) |
| 0.0 |
0.4 |
GO:0045667 |
regulation of osteoblast differentiation(GO:0045667) |
| 0.0 |
0.4 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.2 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.0 |
0.7 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.4 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.9 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.4 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.1 |
GO:1901509 |
regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 |
0.5 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:2000157 |
regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 |
3.0 |
GO:0060021 |
palate development(GO:0060021) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.1 |
GO:0019695 |
choline metabolic process(GO:0019695) |
| 0.0 |
0.5 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.2 |
GO:0042416 |
dopamine biosynthetic process(GO:0042416) |
| 0.0 |
0.4 |
GO:0060259 |
regulation of feeding behavior(GO:0060259) |
| 0.0 |
0.2 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 |
0.7 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.1 |
GO:1902605 |
heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.0 |
0.3 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.0 |
0.1 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.0 |
0.2 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.5 |
GO:0006693 |
prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 |
0.4 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.4 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 |
1.8 |
GO:0006611 |
protein export from nucleus(GO:0006611) |
| 0.0 |
1.3 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.5 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.0 |
0.8 |
GO:0035082 |
axoneme assembly(GO:0035082) |
| 0.0 |
0.3 |
GO:0051703 |
social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 |
1.5 |
GO:0050830 |
defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 |
1.1 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
1.3 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.3 |
GO:0002097 |
tRNA wobble base modification(GO:0002097) |
| 0.0 |
0.5 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.0 |
0.6 |
GO:0042246 |
tissue regeneration(GO:0042246) |
| 0.0 |
0.7 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.0 |
0.2 |
GO:0030825 |
positive regulation of cGMP metabolic process(GO:0030825) positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.4 |
GO:2000010 |
positive regulation of protein localization to cell surface(GO:2000010) |
| 0.0 |
0.3 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.0 |
3.7 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.9 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 |
0.5 |
GO:0042744 |
hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 |
0.2 |
GO:2000637 |
positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 |
0.1 |
GO:0018202 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 |
0.2 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.1 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.0 |
0.8 |
GO:0030177 |
positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.0 |
0.5 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
1.0 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.8 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
1.2 |
GO:0008360 |
regulation of cell shape(GO:0008360) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) response to peptidoglycan(GO:0032494) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.6 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.8 |
GO:0031960 |
response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 |
0.1 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
1.1 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.0 |
GO:1903243 |
negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 |
0.2 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.3 |
GO:0051894 |
positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) |
| 0.0 |
0.4 |
GO:0000002 |
mitochondrial genome maintenance(GO:0000002) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.7 |
GO:0000266 |
mitochondrial fission(GO:0000266) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.7 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.6 |
GO:0045022 |
early endosome to late endosome transport(GO:0045022) |
| 0.0 |
0.1 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.0 |
0.1 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.0 |
0.1 |
GO:0033364 |
mast cell secretory granule organization(GO:0033364) |
| 0.0 |
0.2 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.2 |
GO:0046916 |
cellular iron ion homeostasis(GO:0006879) cellular transition metal ion homeostasis(GO:0046916) |
| 0.0 |
0.1 |
GO:0002484 |
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 |
0.1 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
1.0 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.1 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.0 |
0.0 |
GO:0097300 |
necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 |
0.4 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.1 |
GO:0002002 |
regulation of systemic arterial blood pressure by circulatory renin-angiotensin(GO:0001991) regulation of angiotensin levels in blood(GO:0002002) regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 |
0.2 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.7 |
GO:0006749 |
glutathione metabolic process(GO:0006749) |
| 0.0 |
0.1 |
GO:0030728 |
ovulation(GO:0030728) |
| 0.0 |
0.5 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.3 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.2 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 |
0.1 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.9 |
GO:0071229 |
cellular response to acid chemical(GO:0071229) |
| 0.0 |
0.0 |
GO:0031585 |
regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.0 |
0.3 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.3 |
GO:0042026 |
protein refolding(GO:0042026) |
| 0.0 |
1.0 |
GO:0090090 |
negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 |
0.3 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.3 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.1 |
GO:0043249 |
erythrocyte maturation(GO:0043249) |
| 0.0 |
0.3 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.4 |
GO:0003382 |
epithelial cell morphogenesis(GO:0003382) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.0 |
GO:0031953 |
negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 |
0.3 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |