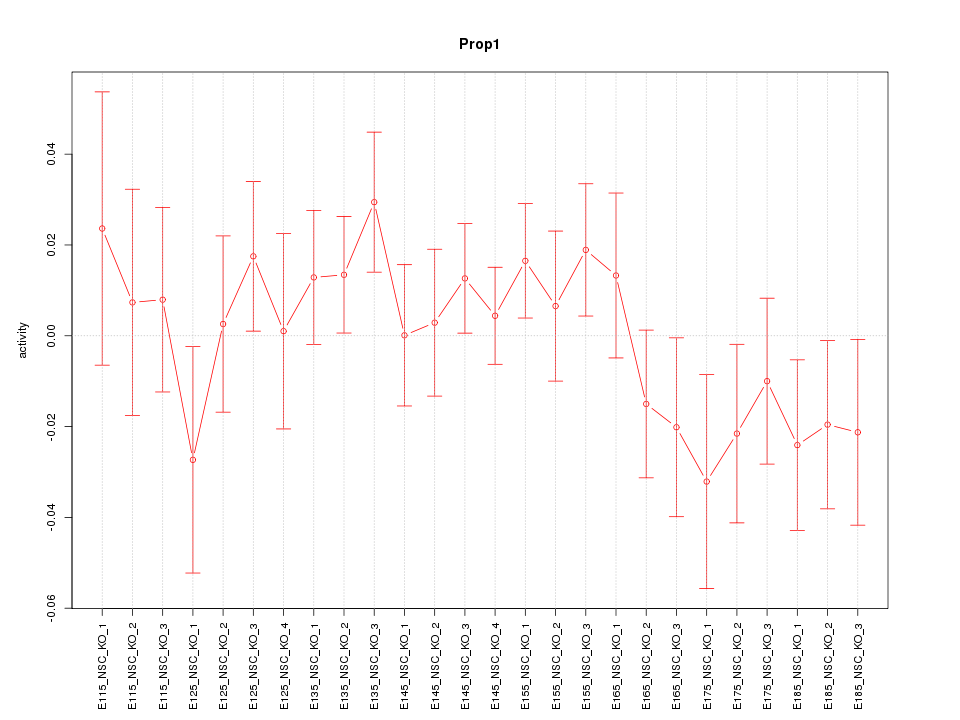
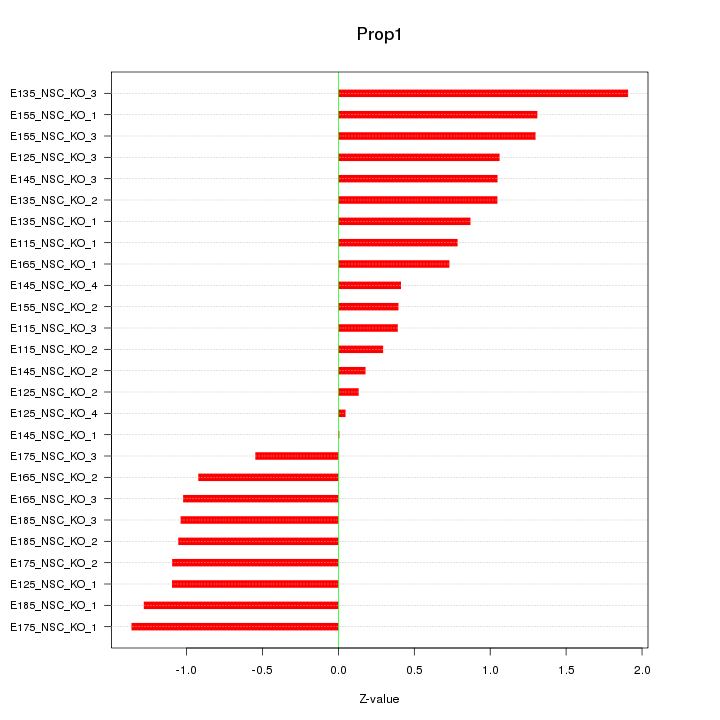
Motif ID: Prop1
Z-value: 0.944
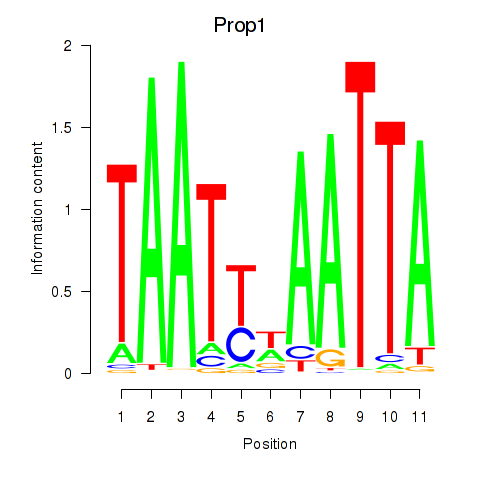
Transcription factors associated with Prop1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Prop1 | ENSMUSG00000044542.3 | Prop1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | mm10_v2_chr11_-_50953745_50953768 | 0.26 | 2.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 | 2.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.8 | 5.6 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.7 | 2.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 | 2.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.5 | 1.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.4 | 1.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 | 1.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.4 | 1.6 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 1.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 0.4 | 3.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 1.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.3 | 1.0 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 1.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.9 | GO:0051595 | response to methylglyoxal(GO:0051595) |
| 0.2 | 0.4 | GO:2000790 | regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.2 | 1.0 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.2 | 1.6 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 0.6 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.2 | 0.5 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.2 | 1.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.3 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.5 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 0.1 | 0.7 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 1.7 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 2.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 5.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.9 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.6 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.3 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.4 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 | 1.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.3 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.1 | 0.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.7 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.1 | 0.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.5 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.0 | 0.6 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.2 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.4 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.3 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:0014891 | striated muscle atrophy(GO:0014891) |
| 0.0 | 0.6 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.3 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.7 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 1.8 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.5 | 1.4 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 1.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 1.8 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 1.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.4 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.7 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 2.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.6 | 5.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 1.1 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 2.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 2.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 0.1 | 0.3 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.5 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 2.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.2 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 1.3 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 0.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) glycerol channel activity(GO:0015254) |
| 0.1 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.1 | GO:0016751 | S-succinyltransferase activity(GO:0016751) |
| 0.1 | 0.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.2 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 1.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.0 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 1.7 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.4 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.9 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 1.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |