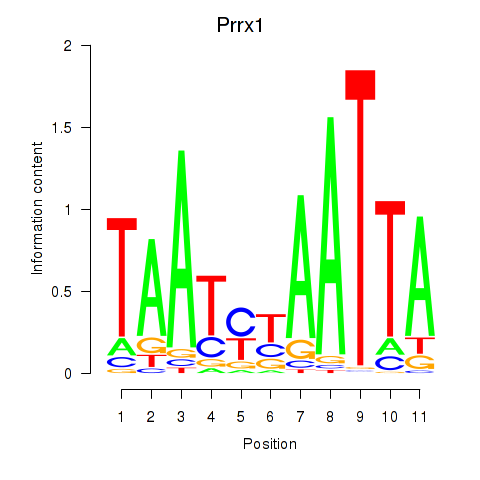
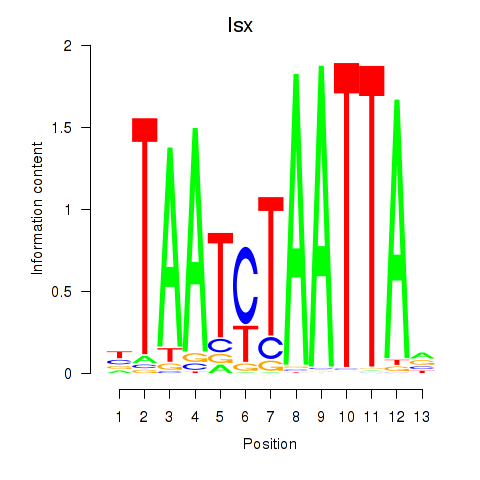
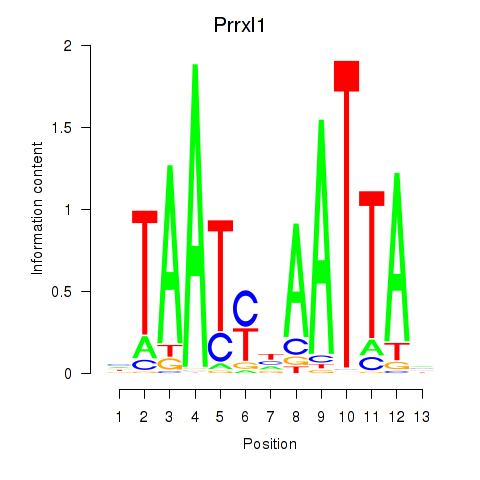
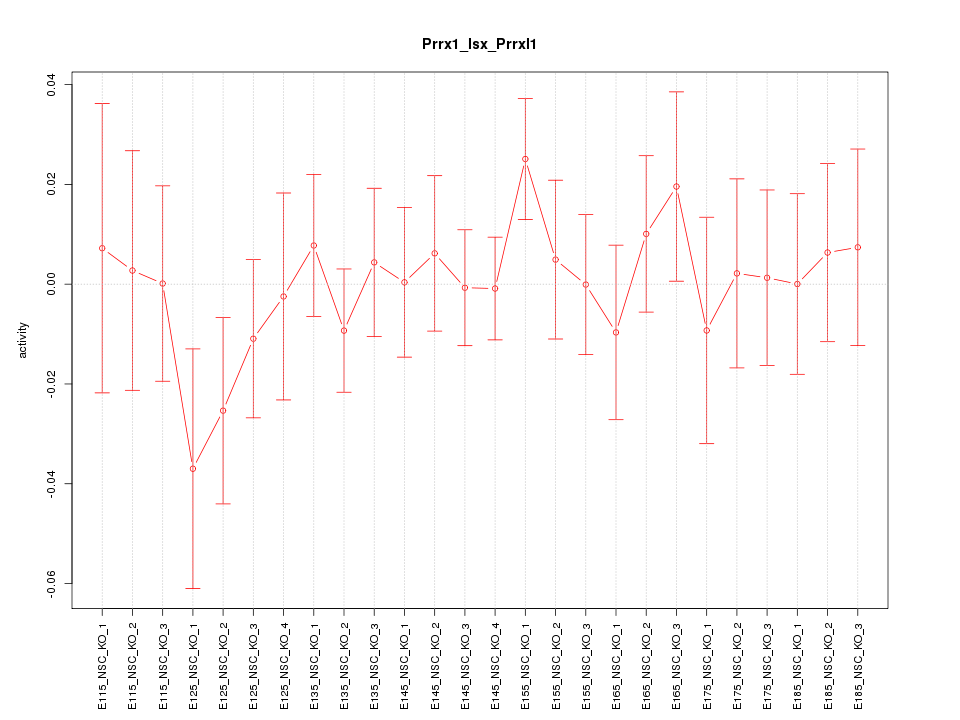
| 1.9 |
5.8 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.6 |
2.3 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 |
2.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.4 |
2.0 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.4 |
1.5 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.2 |
1.0 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
0.7 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.2 |
1.7 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.2 |
0.6 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
1.4 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.2 |
1.9 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.2 |
0.5 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.2 |
0.6 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 |
0.9 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.1 |
1.0 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
0.7 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
1.0 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 |
0.4 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.1 |
0.9 |
GO:0002591 |
positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 |
0.6 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 |
1.1 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.1 |
0.4 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 |
1.7 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.1 |
GO:0002488 |
antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.1 |
0.3 |
GO:0072071 |
apoptotic process involved in heart morphogenesis(GO:0003278) regulation of vascular wound healing(GO:0061043) mesangial cell differentiation(GO:0072007) glomerular mesangial cell differentiation(GO:0072008) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 |
1.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.3 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) |
| 0.1 |
0.3 |
GO:2000670 |
positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 |
0.2 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
0.2 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 |
0.2 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.2 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.1 |
0.3 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.5 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 |
0.2 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.4 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.2 |
GO:0010985 |
negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 |
0.2 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.3 |
GO:0060842 |
arterial endothelial cell differentiation(GO:0060842) |
| 0.1 |
0.2 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 |
0.7 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.1 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
1.0 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 |
0.2 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.3 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.0 |
0.3 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 |
0.2 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:0008582 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 |
0.1 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.0 |
0.1 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.1 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.0 |
0.1 |
GO:0016056 |
rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 |
0.3 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
2.5 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.1 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.4 |
GO:0042428 |
serotonin metabolic process(GO:0042428) |
| 0.0 |
0.1 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 |
0.1 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
0.2 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.5 |
GO:0008206 |
bile acid metabolic process(GO:0008206) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.5 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.1 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 |
0.2 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.9 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.2 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 |
0.2 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 |
0.9 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.1 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
0.1 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
0.1 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 |
0.4 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.0 |
GO:1901856 |
negative regulation of cellular respiration(GO:1901856) |
| 0.0 |
0.1 |
GO:0060126 |
somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.2 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.0 |
0.1 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.0 |
0.4 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.0 |
0.2 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
0.1 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 |
0.1 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
0.2 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.0 |
0.1 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.0 |
0.1 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.0 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 |
0.1 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.1 |
GO:0034285 |
response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 |
0.1 |
GO:0060055 |
angiogenesis involved in wound healing(GO:0060055) |
| 0.0 |
0.1 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
0.1 |
GO:1903772 |
regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.0 |
0.0 |
GO:0035787 |
cell migration involved in kidney development(GO:0035787) |
| 0.0 |
0.1 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 |
0.2 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.1 |
GO:0035791 |
platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 |
0.1 |
GO:0072539 |
T-helper 17 cell differentiation(GO:0072539) |
| 0.0 |
0.0 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 |
0.1 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
1.5 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |